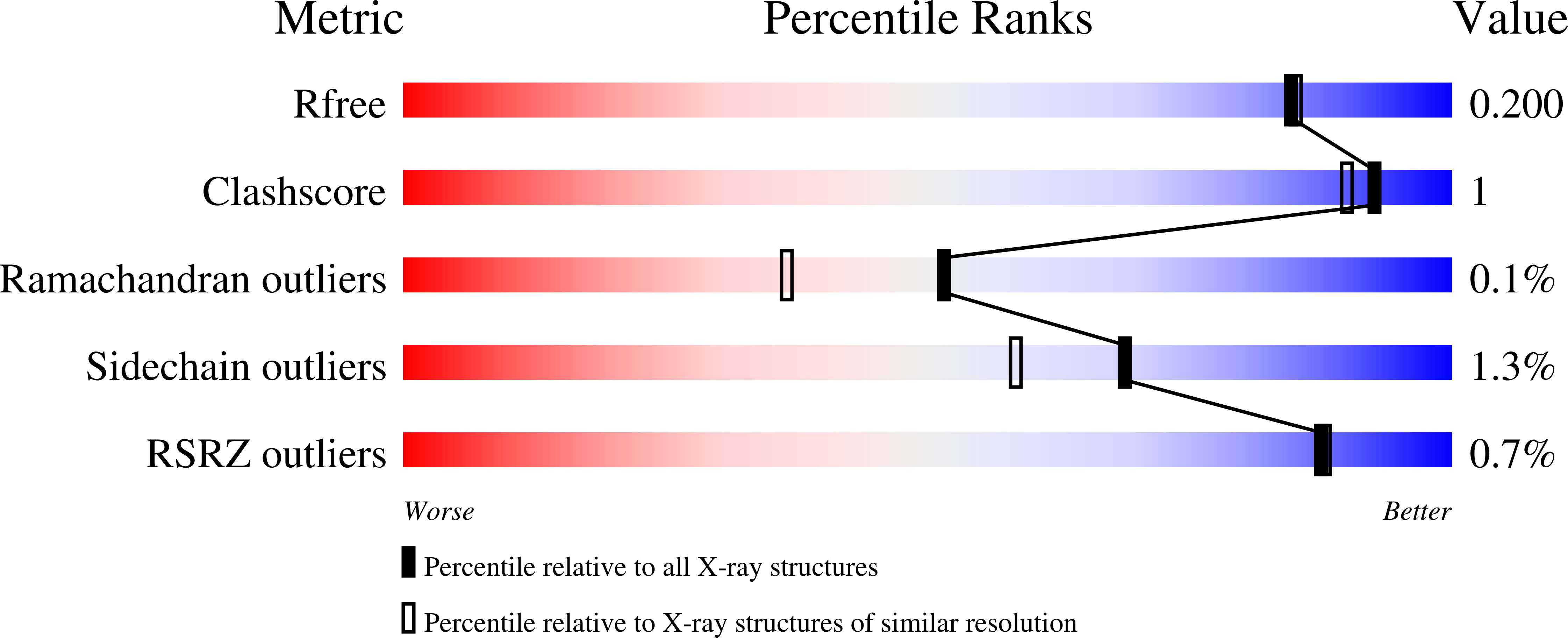
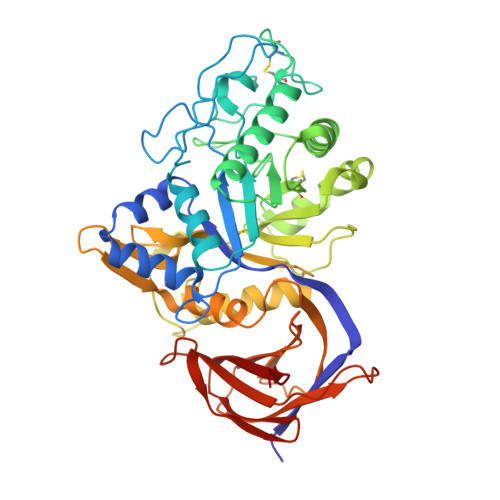
Unique features of the bifunctional GH30 from Thermothelomyces thermophila revealed by structural and mutational studies
Nikolaivits, E., Pentari, C., Kosinas, C., Feiler, C.G., Spiliopoulou, M., Weiss, M.S., Dimarogona, M., Topakas, E.(2021) Carbohydrate Polymers 273: 118553