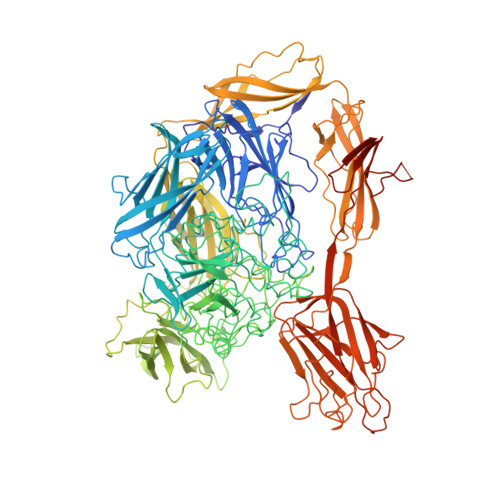
Multitasking in the gut: the X-ray structure of the multidomain BbgIII from Bifidobacterium bifidum offers possible explanations for its alternative functions.
Moroz, O.V., Blagova, E., Lebedev, A.A., Sanchez Rodriguez, F., Rigden, D.J., Tams, J.W., Wilting, R., Vester, J.K., Longhin, E., Hansen, G.H., Krogh, K.B.R.M., Pache, R.A., Davies, G.J., Wilson, K.S.(2021) Acta Crystallogr D Struct Biol 77: 1564-1578
- PubMed: 34866612
- DOI: https://doi.org/10.1107/S2059798321010949
- Primary Citation of Related Structures:
7NIT - PubMed Abstract:
β-Galactosidases catalyse the hydrolysis of lactose into galactose and glucose; as an alternative reaction, some β-galactosidases also catalyse the formation of galactooligosaccharides by transglycosylation. Both reactions have industrial importance: lactose hydrolysis is used to produce lactose-free milk, while galactooligosaccharides have been shown to act as prebiotics. For some multi-domain β-galactosidases, the hydrolysis/transglycosylation ratio can be modified by the truncation of carbohydrate-binding modules. Here, an analysis of BbgIII, a multidomain β-galactosidase from Bifidobacterium bifidum, is presented. The X-ray structure has been determined of an intact protein corresponding to a gene construct of eight domains. The use of evolutionary covariance-based predictions made sequence docking in low-resolution areas of the model spectacularly easy, confirming the relevance of this rapidly developing deep-learning-based technique for model building. The structure revealed two alternative orientations of the CBM32 carbohydrate-binding module relative to the GH2 catalytic domain in the six crystallographically independent chains. In one orientation the CBM32 domain covers the entrance to the active site of the enzyme, while in the other orientation the active site is open, suggesting a possible mechanism for switching between the two activities of the enzyme, namely lactose hydrolysis and transgalactosylation. The location of the carbohydrate-binding site of the CBM32 domain on the opposite site of the module to where it comes into contact with the catalytic GH2 domain is consistent with its involvement in adherence to host cells. The role of the CBM32 domain in switching between hydrolysis and transglycosylation modes offers protein-engineering opportunities for selective β-galactosidase modification for industrial purposes in the future.
Organizational Affiliation:
York Structural Biology Laboratory, Department of Chemistry, University of York, York YO10 5DD, United Kingdom.