Bacterial gasdermins reveal an ancient mechanism of cell death.
Johnson, A.G., Wein, T., Mayer, M.L., Duncan-Lowey, B., Yirmiya, E., Oppenheimer-Shaanan, Y., Amitai, G., Sorek, R., Kranzusch, P.J.(2022) Science 375: 221-225
- PubMed: 35025633
- DOI: https://doi.org/10.1126/science.abj8432
- Primary Citation of Related Structures:
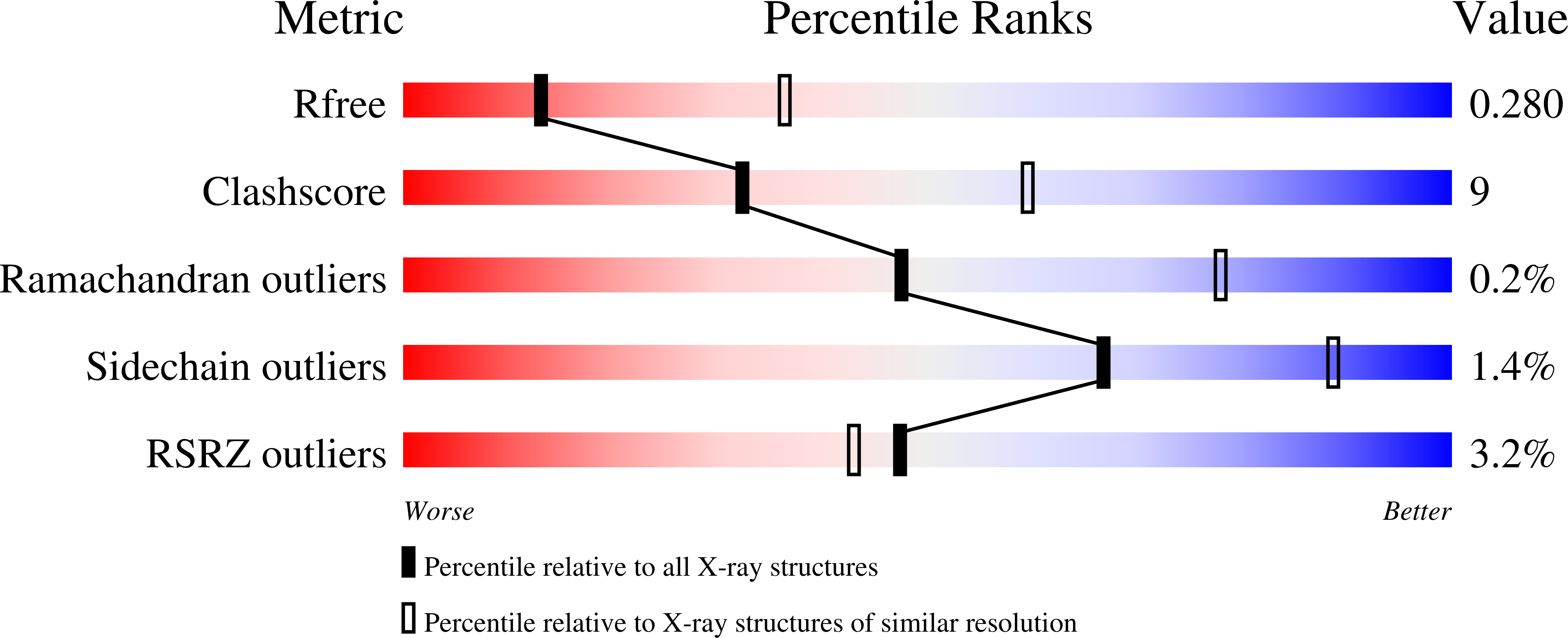
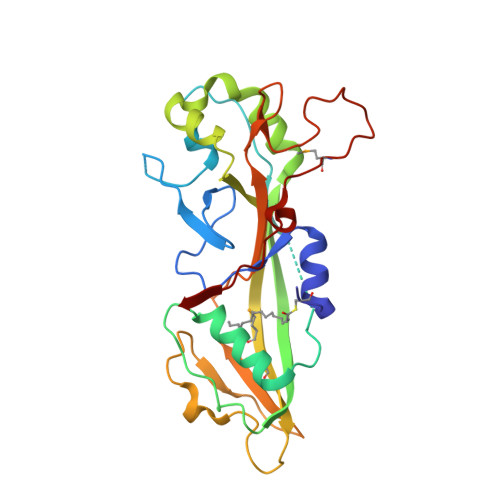
7N50, 7N51, 7N52 - PubMed Abstract:
Gasdermin proteins form large membrane pores in human cells that release immune cytokines and induce lytic cell death. Gasdermin pore formation is triggered by caspase-mediated cleavage during inflammasome signaling and is critical for defense against pathogens and cancer. We discovered gasdermin homologs encoded in bacteria that defended against phages and executed cell death. Structures of bacterial gasdermins revealed a conserved pore-forming domain that was stabilized in the inactive state with a buried lipid modification. Bacterial gasdermins were activated by dedicated caspase-like proteases that catalyzed site-specific cleavage and the removal of an inhibitory C-terminal peptide. Release of autoinhibition induced the assembly of large and heterogeneous pores that disrupted membrane integrity. Thus, pyroptosis is an ancient form of regulated cell death shared between bacteria and animals.
Organizational Affiliation:
Department of Microbiology, Harvard Medical School, Boston, MA 02115, USA.