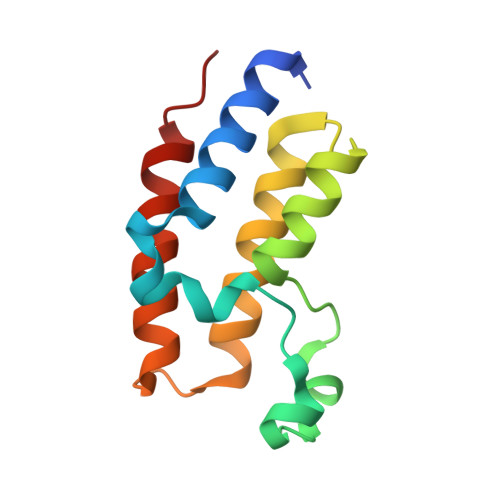
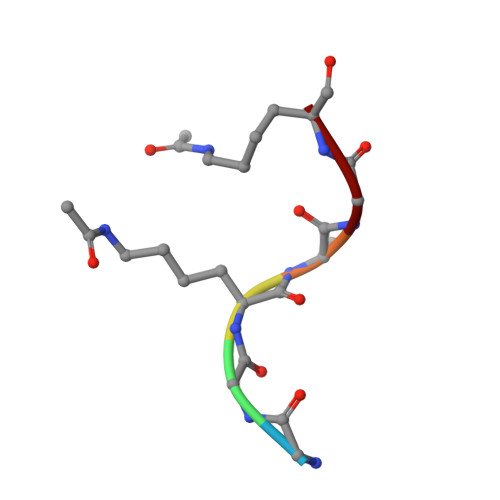
The bromodomains of BET family proteins can recognize diacetylated histone H2A.Z.
Patel, K., Solomon, P.D., Walshe, J.L., Low, J.K.K., Mackay, J.P.(2021) Protein Sci 30: 464-476
- PubMed: 33247496
- DOI: https://doi.org/10.1002/pro.4006
- Primary Citation of Related Structures:
7JX7 - PubMed Abstract:
Chemical modifications of histone tails influence genome accessibility and the transcriptional state of eukaryotic cells. Lysine acetylation is one of the most common modifications and acetyllysine-binding bromodomains (BDs) provide a means for acetyllysine marks to be translated into meaningful cellular responses. Here, we have investigated the mechanism underlying the reported association between the Bromodomain and Extra Terminal (BET) family of BD proteins and the essential histone variant H2A.Z. We use NMR spectroscopy to demonstrate a physical interaction between the N-terminal tail of H2A.Z and the BDs of BRD2, BRD3, and BRD4, and show that the interaction is dependent on lysine acetylation in H2A.Z. The BDs preferentially engage a diacetylated H2A.Z-K4acK7ac motif that is reminiscent of sequences found in other biologically important BET BD target proteins, including histones and transcription factors. A H2A.Z-K7acK11ac motif can also bind BET BDs-with a preference for the second BD of each protein. Chemical shift perturbation mapping of the interactions, together with an X-ray crystal structure of BRD2-BD1 bound to H2A.Z-K4acK7ac, shows that H2A.Z binds the canonical AcK binding pocket of the BDs. This mechanism mirrors the conserved binding mode that is unique to the BET BDs, in which two acetylation marks are read simultaneously by a single BD. Our findings provide structural corroboration of biochemical and cell biological data that link H2A.Z and BET-family proteins, suggesting that the function of H2A.Z is enacted through interactions with these chromatin readers.
Organizational Affiliation:
School of Life and Environmental Sciences, University of Sydney, Sydney, New South Wales, Australia.