Crystallization, structural characterization and kinetic analysis of a GH26 beta-mannanase from Klebsiella oxytoca KUB-CW2-3.
Pongsapipatana, N., Charoenwattanasatien, R., Pramanpol, N., Nguyen, T.H., Haltrich, D., Nitisinprasert, S., Keawsompong, S.(2021) Acta Crystallogr D Struct Biol 77: 1425-1436
- PubMed: 34726170
- DOI: https://doi.org/10.1107/S2059798321009992
- Primary Citation of Related Structures:
7EET - PubMed Abstract:
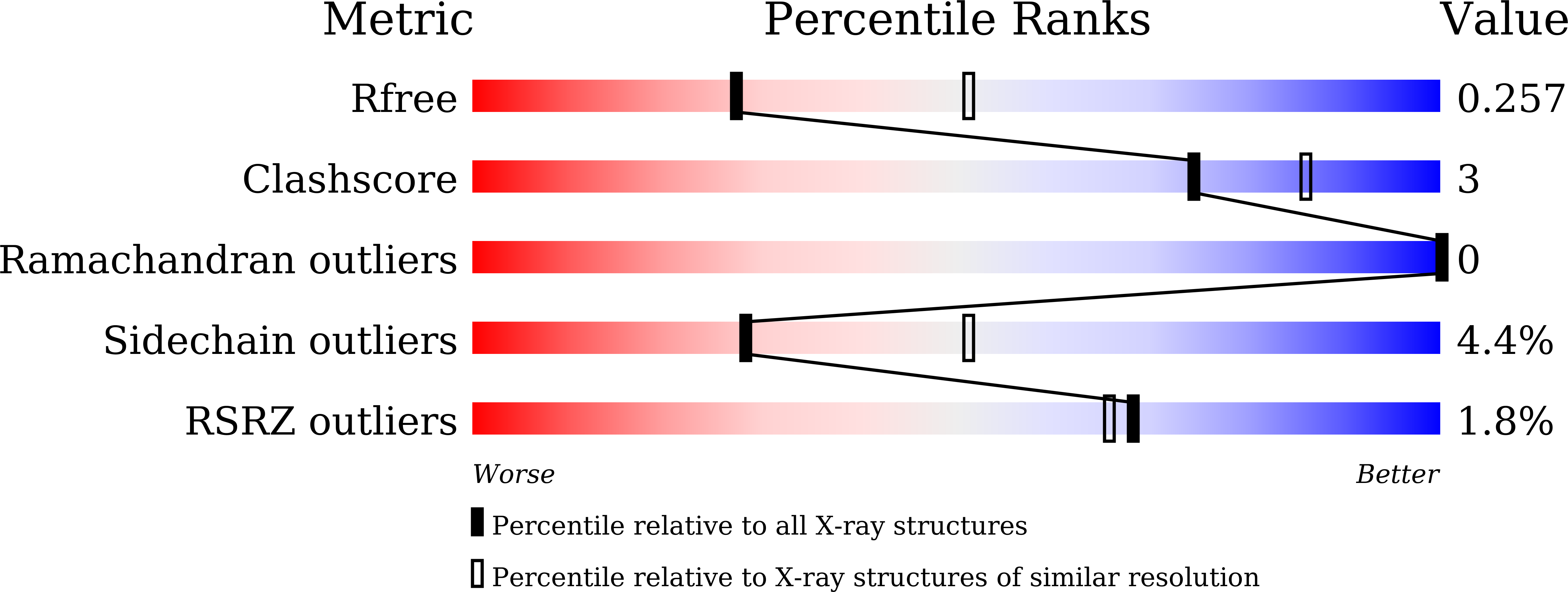
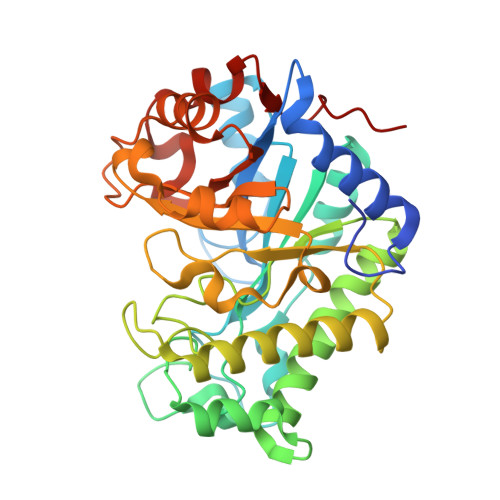
β-Mannanase (EC 3.2.1.78) is an enzyme that cleaves within the backbone of mannan-based polysaccharides at β-1,4-linked D-mannose residues, resulting in the formation of mannooligosaccharides (MOS), which are potential prebiotics. The GH26 β-mannanase KMAN from Klebsiella oxytoca KUB-CW2-3 shares 49-72% amino-acid sequence similarity with β-mannanases from other sources. The crystal structure of KMAN at a resolution of 2.57 Å revealed an open cleft-shaped active site. The enzyme structure is based on a (β/α) 8 -barrel architecture, which is a typical characteristic of clan A glycoside hydrolase enzymes. The putative catalytic residues Glu183 and Glu282 are located on the loop connected to β-strand 4 and at the end of β-strand 7, respectively. KMAN digests linear MOS with a degree of polymerization (DP) of between 4 and 6, with high catalytic efficiency (k cat /K m ) towards DP6 (2571.26 min -1 mM -1 ). The predominant end products from the hydrolysis of locust bean gum, konjac glucomannan and linear MOS are mannobiose and mannotriose. It was observed that KMAN requires at least four binding sites for the binding of substrate molecules and hydrolysis. Molecular docking of mannotriose and galactosyl-mannotetraose to KMAN confirmed its mode of action, which prefers linear substrates to branched substrates.
Organizational Affiliation:
Specialized Research Unit: Prebiotics and Probiotics for Health, Department of Biotechnology, Faculty of Agro-Industry, Kasetsart University, Bangkok 10900, Thailand.