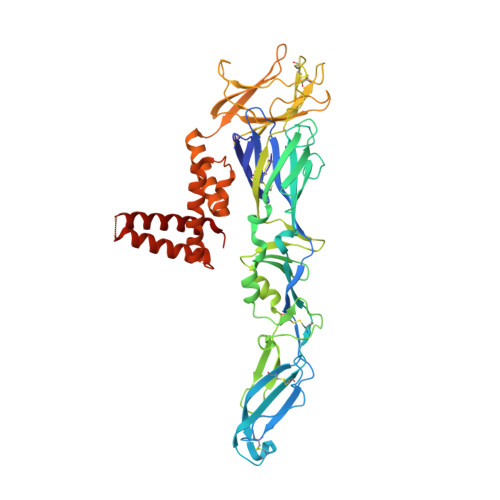
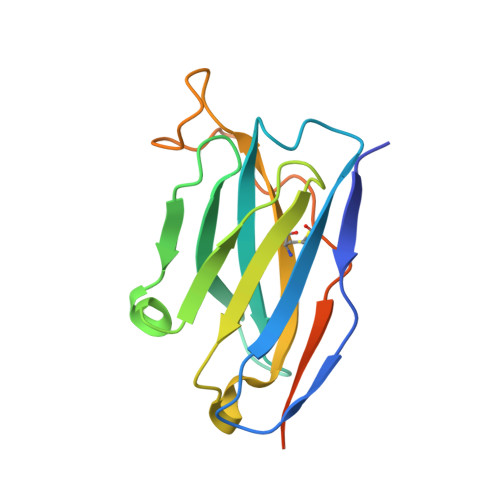
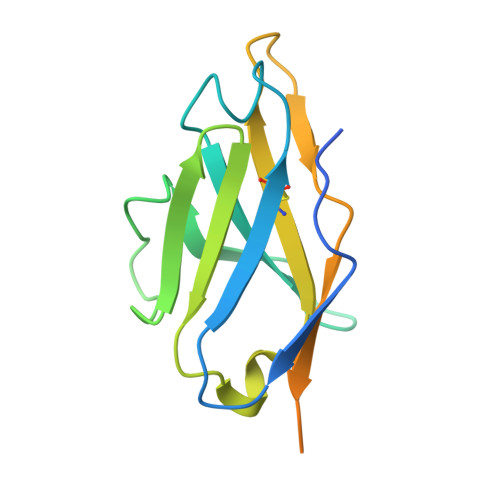
The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Sharma, A., Zhang, X., Dejnirattisai, W., Dai, X., Gong, D., Wongwiwat, W., Duquerroy, S., Rouvinski, A., Vaney, M.C., Guardado-Calvo, P., Haouz, A., England, P., Sun, R., Zhou, Z.H., Mongkolsapaya, J., Screaton, G.R., Rey, F.A.(2021) Cell 184: 6052
- PubMed: 34852239
- DOI: https://doi.org/10.1016/j.cell.2021.11.010
- Primary Citation of Related Structures:
7A3N, 7A3O, 7A3P, 7A3Q, 7A3R, 7A3S, 7A3T, 7A3U, 7CTH - PubMed Abstract:
The human monoclonal antibody C10 exhibits extraordinary cross-reactivity, potently neutralizing Zika virus (ZIKV) and the four serotypes of dengue virus (DENV1-DENV4). Here we describe a comparative structure-function analysis of C10 bound to the envelope (E) protein dimers of the five viruses it neutralizes. We demonstrate that the C10 Fab has high affinity for ZIKV and DENV1 but not for DENV2, DENV3, and DENV4. We further show that the C10 interaction with the latter viruses requires an E protein conformational landscape that limits binding to only one of the three independent epitopes per virion. This limited affinity is nevertheless counterbalanced by the particle's icosahedral organization, which allows two different dimers to be reached by both Fab arms of a C10 immunoglobulin. The epitopes' geometric distribution thus confers C10 its exceptional neutralization breadth. Our results highlight the importance not only of paratope/epitope complementarity but also the topological distribution for epitope-focused vaccine design.
Organizational Affiliation:
Institut Pasteur, Université de Paris, CNRS UMR3569, Unité de Virologie Structurale, 75015 Paris, France.