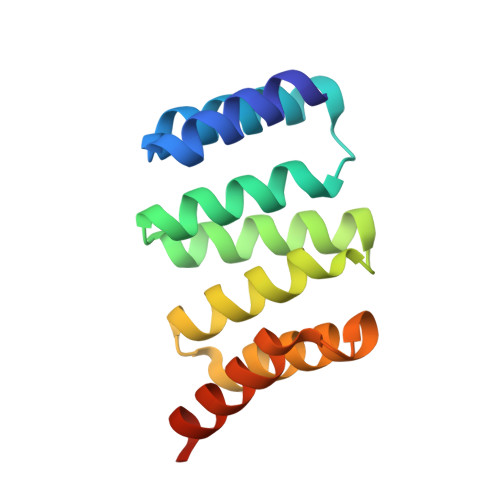
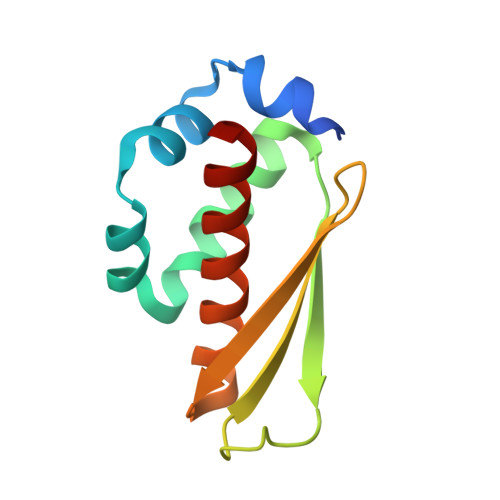
The interaction between RPAP3 and TRBP reveals a possible involvement of the HSP90/R2TP chaperone complex in the regulation of miRNA activity.
Abel, Y., Charron, C., Virciglio, C., Bourguignon-Igel, V., Quinternet, M., Chagot, M.E., Robert, M.C., Verheggen, C., Branlant, C., Bertrand, E., Manival, X., Charpentier, B., Rederstorff, M.(2022) Nucleic Acids Res 50: 2172-2189
- PubMed: 35150569
- DOI: https://doi.org/10.1093/nar/gkac086
- Primary Citation of Related Structures:
6ZBK - PubMed Abstract:
MicroRNAs silence mRNAs by guiding the RISC complex. RISC assembly occurs following cleavage of pre-miRNAs by Dicer, assisted by TRBP or PACT, and the transfer of miRNAs to AGO proteins. The R2TP complex is an HSP90 co-chaperone involved in the assembly of ribonucleoprotein particles. Here, we show that the R2TP component RPAP3 binds TRBP but not PACT. The RPAP3-TPR1 domain interacts with the TRBP-dsRBD3, and the 1.5 Å resolution crystal structure of this complex identifies key residues involved in the interaction. Remarkably, binding of TRBP to RPAP3 or Dicer is mutually exclusive. Additionally, we found that AGO(1/2), TRBP and Dicer are all sensitive to HSP90 inhibition, and that TRBP sensitivity is increased in the absence of RPAP3. Finally, RPAP3 seems to impede miRNA activity, raising the possibility that the R2TP chaperone might sequester TRBP to regulate the miRNA pathway.
Organizational Affiliation:
Université de Lorraine, CNRS, IMoPA, F-54000 Nancy, France.