Computationally designed dual-color MRI reporters for noninvasive imaging of transgene expression.
Allouche-Arnon, H., Khersonsky, O., Tirukoti, N.D., Peleg, Y., Dym, O., Albeck, S., Brandis, A., Mehlman, T., Avram, L., Harris, T., Yadav, N.N., Fleishman, S.J., Bar-Shir, A.(2022) Nat Biotechnol 40: 1143-1149
- PubMed: 35102291
- DOI: https://doi.org/10.1038/s41587-021-01162-5
- Primary Citation of Related Structures:
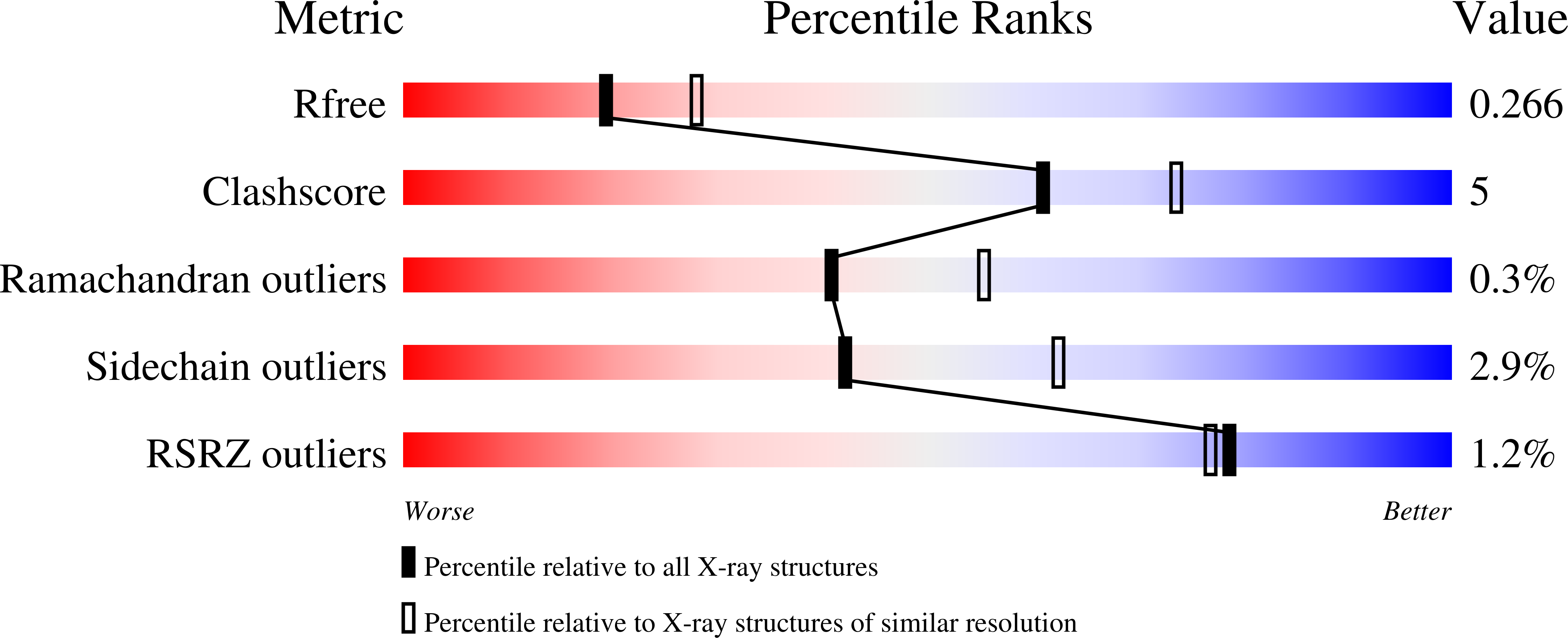
6YBH - PubMed Abstract:
Imaging of gene-expression patterns in live animals is difficult to achieve with fluorescent proteins because tissues are opaque to visible light. Imaging of transgene expression with magnetic resonance imaging (MRI), which penetrates to deep tissues, has been limited by single reporter visualization capabilities. Moreover, the low-throughput capacity of MRI limits large-scale mutagenesis strategies to improve existing reporters. Here we develop an MRI system, called GeneREFORM, comprising orthogonal reporters for two-color imaging of transgene expression in deep tissues. Starting from two promiscuous deoxyribonucleoside kinases, we computationally designed highly active, orthogonal enzymes ('reporter genes') that specifically phosphorylate two MRI-detectable synthetic deoxyribonucleosides ('reporter probes'). Systemically administered reporter probes exclusively accumulate in cells expressing the designed reporter genes, and their distribution is displayed as pseudo-colored MRI maps based on dynamic proton exchange for noninvasive visualization of transgene expression. We envision that future extensions of GeneREFORM will pave the way to multiplexed deep-tissue mapping of gene expression in live animals.
Organizational Affiliation:
Department of Molecular Chemistry and Materials Science, Weizmann Institute of Science, Rehovot, Israel.