Challenging a Preconception: Optoacoustic Spectrum Differs from the Optical Absorption Spectrum of Proteins and Dyes for Molecular Imaging.
Fuenzalida Werner, J.P., Huang, Y., Mishra, K., Janowski, R., Vetschera, P., Heichler, C., Chmyrov, A., Neufert, C., Niessing, D., Ntziachristos, V., Stiel, A.C.(2020) Anal Chem 92: 10717-10724
- PubMed: 32640156
- DOI: https://doi.org/10.1021/acs.analchem.0c01902
- Primary Citation of Related Structures:
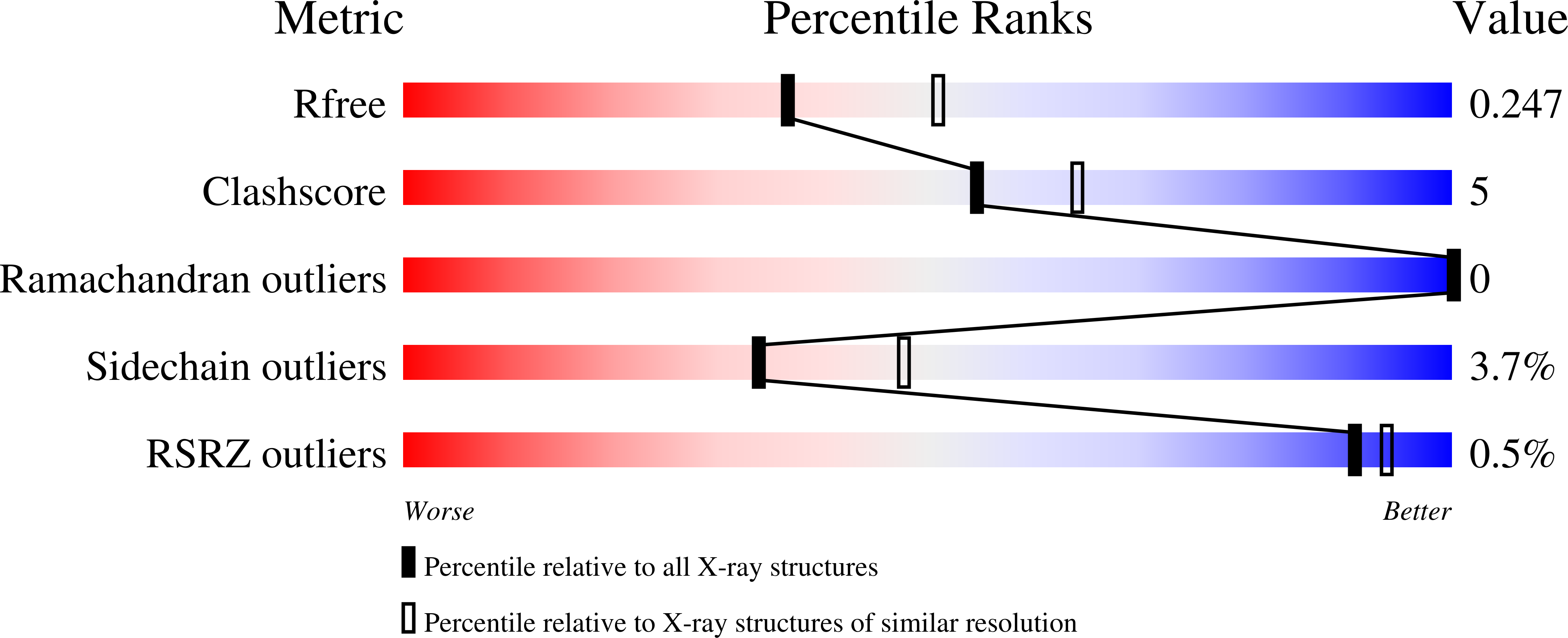
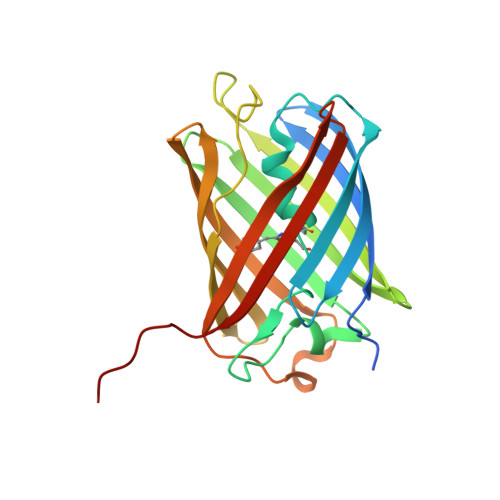
6Y1G - PubMed Abstract:
Optoacoustic (photoacoustic) imaging has seen marked advances in detection and data analysis, but there is less progress in understanding the photophysics of common optoacoustic contrast agents. This gap blocks the development of novel agents and the accurate analysis and interpretation of multispectral optoacoustic images. To close it, we developed a multimodal laser spectrometer (MLS) to enable the simultaneous measurement of optoacoustic, absorbance, and fluorescence spectra. Herein, we employ MLS to analyze contrast agents (methylene blue, rhodamine 800, Alexa Fluor 750, IRDye 800CW, and indocyanine green) and proteins (sfGFP, mCherry, mKate, HcRed, iRFP720, and smURFP). We found that the optical absorption spectrum does not correlate with the optoacoustic spectrum for the majority of the analytes. We determined that for dyes, the transition underlying an aggregation state has more optoacoustic signal generation efficiency than the monomer transition. For proteins we found a favored optoacoustic relaxation that stems from the neutral or zwitterionic chromophores and unreported photoswitching behavior of tdTomato and HcRed. We then crystalized HcRed in its photoswitch optoacoustic state, confirming structurally the change in isomerization with respect to HcReds' fluorescence state. Finally, on the example of the widely used label tdTomato and the dye indocyanine green, we show the importance of correct photophysical (e.g., spectral and kinetic) information as a prerequisite for spectral-unmixing for in vivo imaging.
Organizational Affiliation:
Institute of Biological and Medical Imaging (IBMI), Helmholtz Zentrum München, D-85764 Neuherberg, Germany.