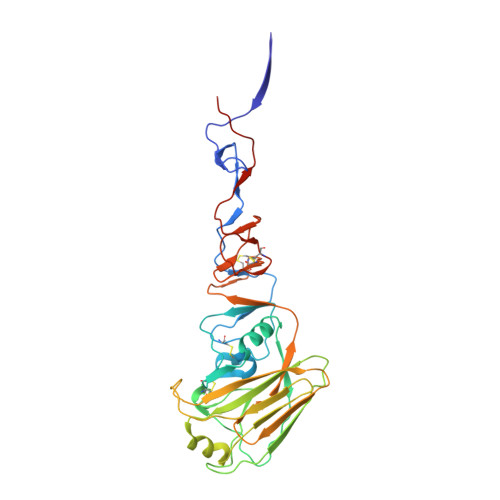
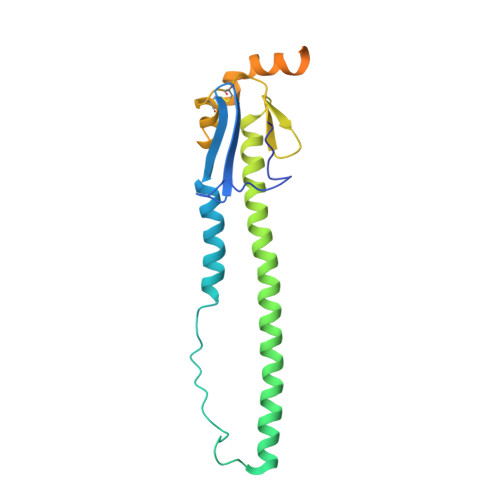
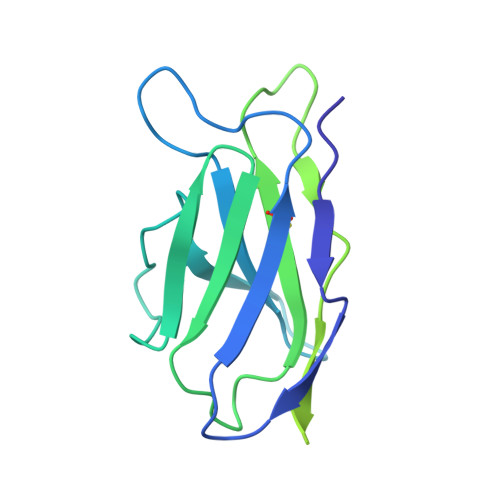
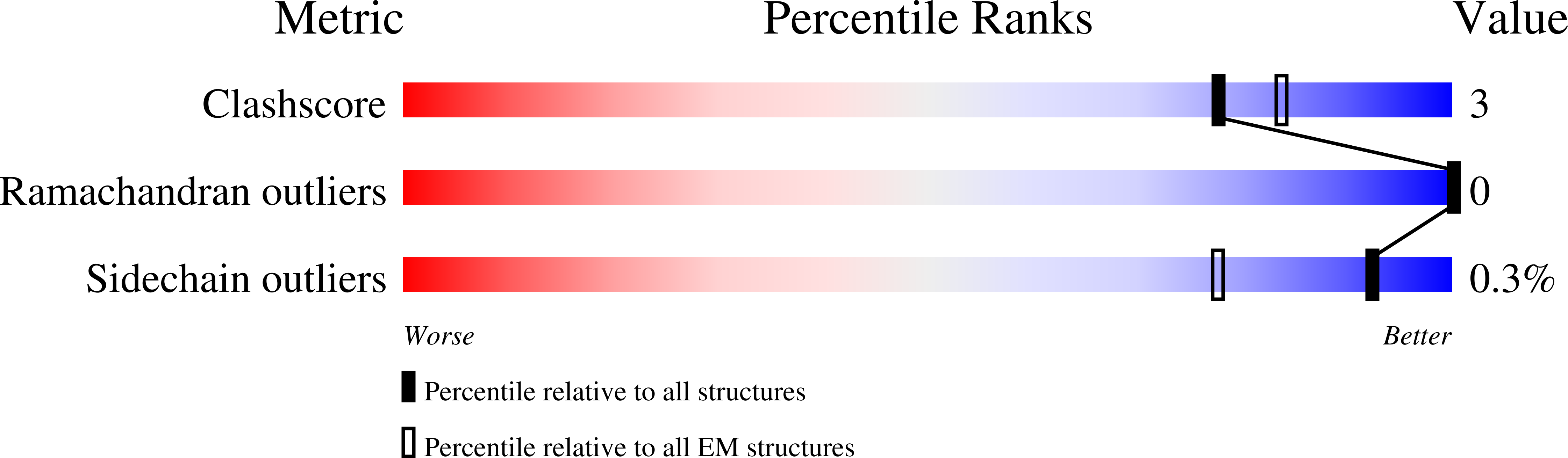Structure of an influenza group 2-neutralizing antibody targeting the hemagglutinin stem supersite.
Cheung, C.S., Gorman, J., Andrews, S.F., Rawi, R., Reveiz, M., Shen, C.H., Wang, Y., Harris, D.R., Nazzari, A.F., Olia, A.S., Raab, J., Teng, I.T., Verardi, R., Wang, S., Yang, Y., Chuang, G.Y., McDermott, A.B., Zhou, T., Kwong, P.D.(2022) Structure
- PubMed: 35489332
- DOI: https://doi.org/10.1016/j.str.2022.04.003
- Primary Citation of Related Structures:
6WXL - PubMed Abstract:
Several influenza antibodies with broad group 2 neutralization have recently been isolated. Here, we analyze the structure, class, and binding of one of these antibodies from an H7N9 vaccine trial, 315-19-1D12. The cryo-EM structure of 315-19-1D12 Fab in complex with the hemagglutinin (HA) trimer revealed the antibody to recognize the helix A region of the HA stem, at the supersite of vulnerability recognized by group 1-specific and by cross-group-neutralizing antibodies. 315-19-1D12 was derived from HV1-2 and KV2-28 genes and appeared to form a new antibody class. Bioinformatic analysis indicated its group 2 neutralization specificity to be a consequence of four key residue positions. We specifically tested the impact of the group 1-specific N33 glycan, which decreased but did not abolish group 2 binding of 315-19-1D12. Overall, this study highlights the recognition of a broad group 2-neutralizing antibody, revealing unexpected diversity in neutralization specificity for antibodies that recognize the HA stem supersite.
Organizational Affiliation:
Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.