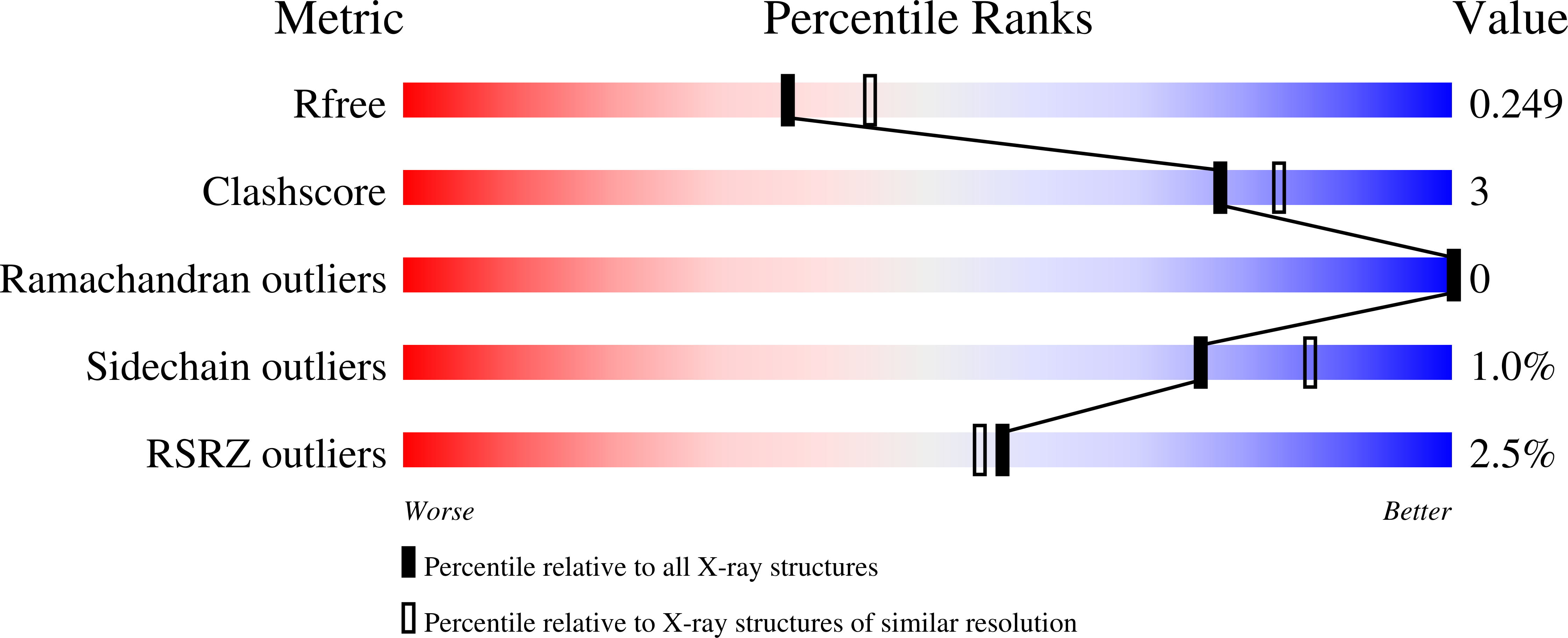
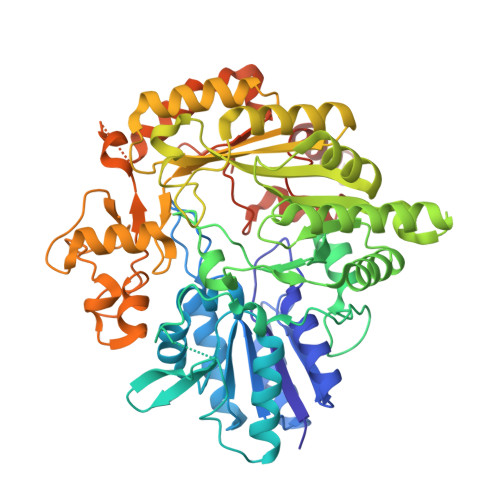
Structural basis for non-radical catalysis by TsrM, a radical SAM methylase.
Knox, H.L., Chen, P.Y., Blaszczyk, A.J., Mukherjee, A., Grove, T.L., Schwalm, E.L., Wang, B., Drennan, C.L., Booker, S.J.(2021) Nat Chem Biol 17: 485-491
- PubMed: 33462497
- DOI: https://doi.org/10.1038/s41589-020-00717-y
- Primary Citation of Related Structures:
6WTE, 6WTF - PubMed Abstract:
Tryptophan 2C methyltransferase (TsrM) methylates C2 of the indole ring of L-tryptophan during biosynthesis of the quinaldic acid moiety of thiostrepton. TsrM is annotated as a cobalamin-dependent radical S-adenosylmethionine (SAM) methylase; however, TsrM does not reductively cleave SAM to the universal 5'-deoxyadenosyl 5'-radical intermediate, a hallmark of radical SAM (RS) enzymes. Herein, we report structures of TsrM from Kitasatospora setae, which are the first structures of a cobalamin-dependent radical SAM methylase. Unexpectedly, the structures show an essential arginine residue that resides in the proximal coordination sphere of the cobalamin cofactor, and a [4Fe-4S] cluster that is ligated by a glutamyl residue and three cysteines in a canonical CXXXCXXC RS motif. Structures in the presence of substrates suggest a substrate-assisted mechanism of catalysis, wherein the carboxylate group of SAM serves as a general base to deprotonate N1 of the tryptophan substrate, facilitating the formation of a C2 carbanion.
Organizational Affiliation:
Department of Chemistry, Pennsylvania State University, University Park, PA, USA.