Novel functional insights into a modified sugar-binding protein from Synechococcus MITS9220.
Ford, B.A., Michie, K.A., Paulsen, I.T., Mabbutt, B.C., Shah, B.S.(2022) Sci Rep 12: 4805-4805
- PubMed: 35314715
- DOI: https://doi.org/10.1038/s41598-022-08459-8
- Primary Citation of Related Structures:
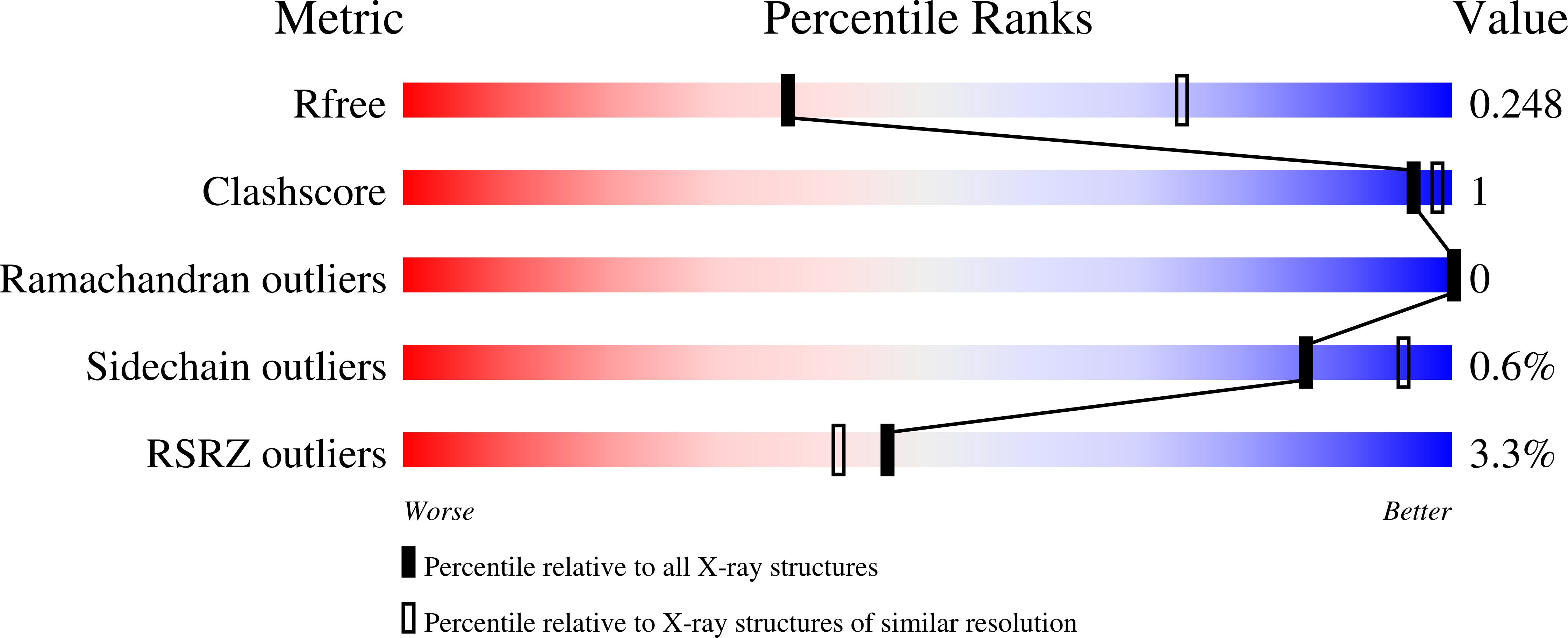
6WPM, 6WPN - PubMed Abstract:
Paradigms of metabolic strategies employed by photoautotrophic marine picocyanobacteria have been challenged in recent years. Based on genomic annotations, picocyanobacteria are predicted to assimilate organic nutrients via ATP-binding cassette importers, a process mediated by substrate-binding proteins. We report the functional characterisation of a modified sugar-binding protein, MsBP, from a marine Synechococcus strain, MITS9220. Ligand screening of MsBP shows a specific affinity for zinc (K D ~ 1.3 μM) and a preference for phosphate-modified sugars, such as fructose-1,6-biphosphate, in the presence of zinc (K D ~ 5.8 μM). Our crystal structures of apo MsBP (no zinc or substrate-bound) and Zn-MsBP (with zinc-bound) show that the presence of zinc induces structural differences, leading to a partially-closed substrate-binding cavity. The Zn-MsBP structure also sequesters several sulphate ions from the crystallisation condition, including two in the binding cleft, appropriately placed to mimic the orientation of adducts of a biphosphate hexose. Combined with a previously unseen positively charged binding cleft in our two structures and our binding affinity data, these observations highlight novel molecular variations on the sugar-binding SBP scaffold. Our findings lend further evidence to a proposed sugar acquisition mechanism in picocyanobacteria alluding to a mixotrophic strategy within these ubiquitous photosynthetic bacteria.
Organizational Affiliation:
School of Natural Sciences, Macquarie University, Sydney, Australia.