Residual Structure in the Denatured State of the Fast-Folding UBA(1) Domain from the Human DNA Excision Repair Protein HHR23A.
Becht, D.C., Leavens, M.J., Zeng, B., Rothfuss, M.T., Briknarova, K., Bowler, B.E.(2022) Biochemistry 61: 767-784
- PubMed: 35430812
- DOI: https://doi.org/10.1021/acs.biochem.2c00011
- Primary Citation of Related Structures:
6W2H - PubMed Abstract:
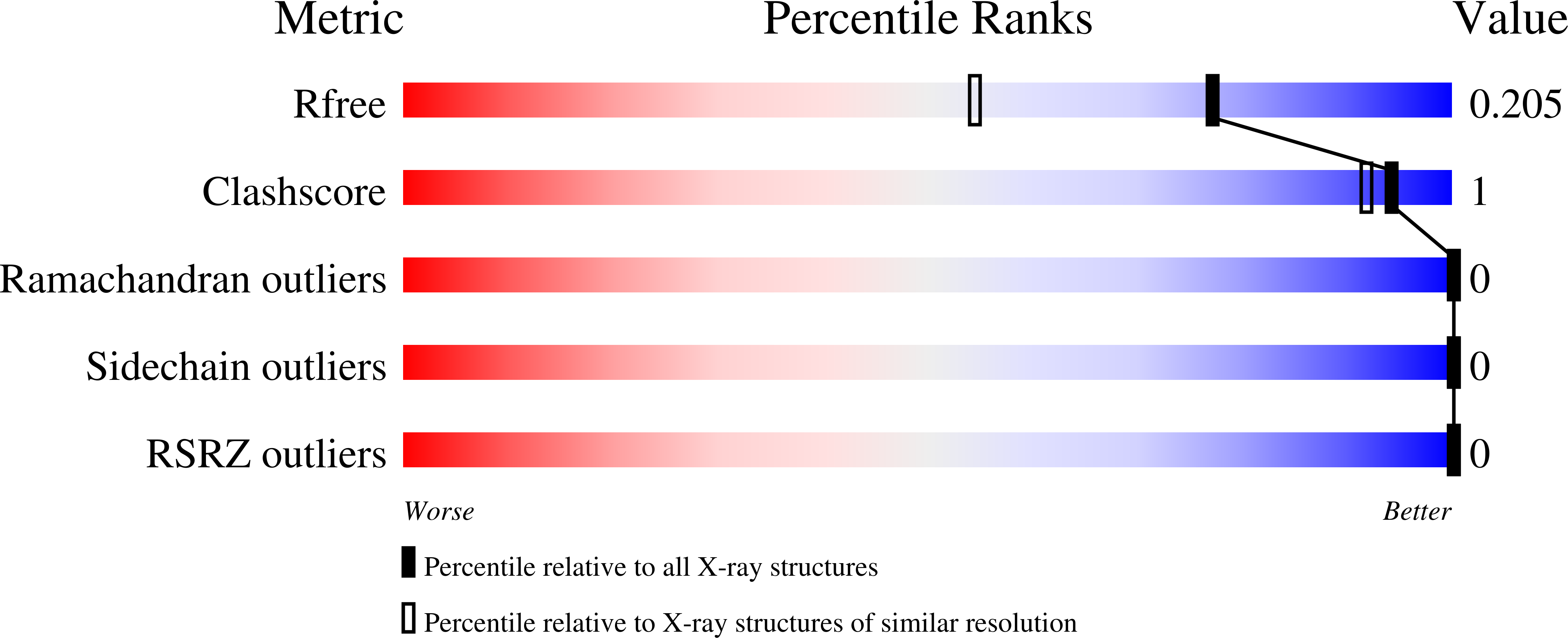
The structure of the first ubiquitin-associated domain from HHR23A, UBA(1), was determined by X-ray crystallography at a 1.60 Å resolution, and its stability, folding kinetics, and residual structure under denaturing conditions have been investigated. The concentration dependence of thermal denaturation and size-exclusion chromatography indicate that UBA(1) is monomeric. Guanidine hydrochloride (GdnHCl) denaturation experiments reveal that the unfolding free energy, Δ G u °'(H 2 O), of UBA(1) is 2.4 kcal mol -1 . Stopped-flow folding kinetics indicates sub-millisecond folding with only proline isomerization phases detectable at 25 °C. The full folding kinetics are observable at 4 °C, yielding a folding rate constant, k f , in the absence of a denaturant of 13,000 s -1 and a Tanford β-value of 0.80, consistent with a compact transition state. Evaluation of the secondary structure via circular dichroism shows that the residual helical structure in the denatured state is replaced by polyproline II structure as the GdnHCl concentration increases. Analysis of NMR secondary chemical shifts for backbone 15 NH, 13 CO, and 13 Cα atoms between 4 and 7 M GdnHCl shows three islands of residual helical secondary structure that align in sequence with the three native-state helices. Extrapolation of the NMR data to 0 M GdnHCl demonstrates that helical structure would populate to 17-33% in the denatured state under folding conditions. Comparison with NMR data for a peptide corresponding to helix 1 indicates that this helix is stabilized by transient tertiary interactions in the denatured state of UBA(1). The high helical content in the denatured state, which is enhanced by transient tertiary interactions, suggests a diffusion-collision folding mechanism.
Organizational Affiliation:
Department of Chemistry & Biochemistry, University of Montana, Missoula, Montana 59812, United States.