Potent Henipavirus Neutralization by Antibodies Recognizing Diverse Sites on Hendra and Nipah Virus Receptor Binding Protein.
Dong, J., Cross, R.W., Doyle, M.P., Kose, N., Mousa, J.J., Annand, E.J., Borisevich, V., Agans, K.N., Sutton, R., Nargi, R., Majedi, M., Fenton, K.A., Reichard, W., Bombardi, R.G., Geisbert, T.W., Crowe Jr., J.E.(2020) Cell 183: 1536-1550.e17
- PubMed: 33306954
- DOI: https://doi.org/10.1016/j.cell.2020.11.023
- Primary Citation of Related Structures:
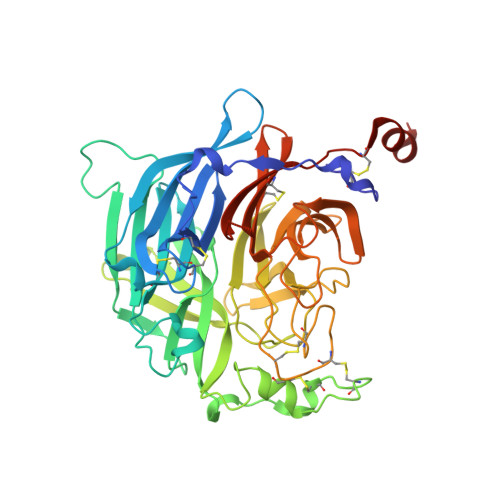
6VY4, 6VY5, 6VY6 - PubMed Abstract:
Hendra (HeV) and Nipah (NiV) viruses are emerging zoonotic pathogens in the Henipavirus genus causing outbreaks of disease with very high case fatality rates. Here, we report the first naturally occurring human monoclonal antibodies (mAbs) against HeV receptor binding protein (RBP). All isolated mAbs neutralized HeV, and some also neutralized NiV. Epitope binning experiments identified five major antigenic sites on HeV-RBP. Animal studies demonstrated that the most potent cross-reactive neutralizing mAbs, HENV-26 and HENV-32, protected ferrets in lethal models of infection with NiV Bangladesh 3 days after exposure. We solved the crystal structures of mAb HENV-26 in complex with both HeV-RBP and NiV-RBP and of mAb HENV-32 in complex with HeV-RBP. The studies reveal diverse sites of vulnerability on RBP recognized by potent human mAbs that inhibit virus by multiple mechanisms. These studies identify promising prophylactic antibodies and define protective epitopes that can be used in rational vaccine design.
Organizational Affiliation:
The Vanderbilt Vaccine Center, Vanderbilt University Medical Center, Nashville, TN 37232, USA.