NKNK: a New Essential Motif in the C-Terminal Domain of HIV-1 Group M Integrases.
Kanja, M., Cappy, P., Levy, N., Oladosu, O., Schmidt, S., Rossolillo, P., Winter, F., Gasser, R., Moog, C., Ruff, M., Negroni, M., Lener, D.(2020) J Virol 94
- PubMed: 32727879
- DOI: https://doi.org/10.1128/JVI.01035-20
- Primary Citation of Related Structures:
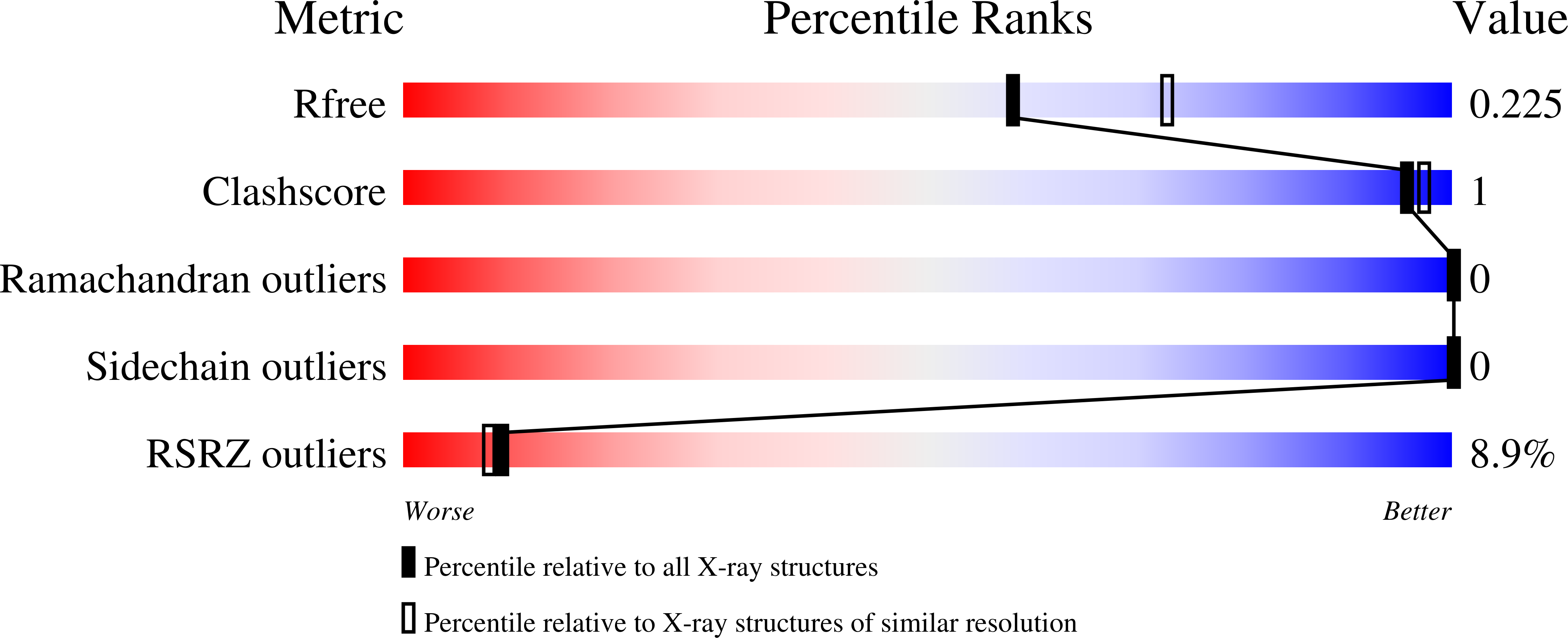
6T6E, 6T6I, 6T6J - PubMed Abstract:
Using coevolution network interference based on comparison of two phylogenetically distantly related isolates, one from the main group M and the other from the minor group O of HIV-1, we identify, in the C-terminal domain (CTD) of integrase, a new functional motif constituted by four noncontiguous amino acids (N 222 K 240 N 254 K 273 ). Mutating the lysines abolishes integration through decreased 3' processing and inefficient nuclear import of reverse-transcribed genomes. Solution of the crystal structures of wild-type (wt) and mutated CTDs shows that the motif generates a positive surface potential that is important for integration. The number of charges in the motif appears more crucial than their position within the motif. Indeed, the positions of the K's could be permutated or additional K's could be inserted in the motif, generally without affecting integration per se Despite this potential genetic flexibility, the NKNK arrangement is strictly conserved in natural sequences, indicative of an effective purifying selection exerted at steps other than integration. Accordingly, reverse transcription was reduced even in the mutants that retained wt integration levels, indicating that specifically the wt sequence is optimal for carrying out the multiple functions that integrase exerts. We propose that the existence of several amino acid arrangements within the motif, with comparable efficiencies of integration per se , might have constituted an asset for the acquisition of additional functions during viral evolution. IMPORTANCE Intensive studies of HIV-1 have revealed its extraordinary ability to adapt to environmental and immunological challenges, an ability that is also at the basis of antiviral treatment escape. Here, by deconvoluting the different roles of the viral integrase in the various steps of the infectious cycle, we report how the existence of alternative equally efficient structural arrangements for carrying out one function opens up the possibility of adapting to the optimization of further functionalities exerted by the same protein. Such a property provides an asset to increase the efficiency of the infectious process. On the other hand, though, the identification of this new motif provides a potential target for interfering simultaneously with multiple functions of the protein.
Organizational Affiliation:
Université de Strasbourg, CNRS, Architecture et Réactivité de l'ARN, Strasbourg, France.