Conformational editing of intrinsically disordered protein by alpha-methylation.
Bauer, V., Schmidtgall, B., Gogl, G., Dolenc, J., Osz, J., Nomine, Y., Kostmann, C., Cousido-Siah, A., Mitschler, A., Rochel, N., Trave, G., Kieffer, B., Torbeev, V.(2020) Chem Sci 12: 1080-1089
- PubMed: 34163874
- DOI: https://doi.org/10.1039/d0sc04482b
- Primary Citation of Related Structures:
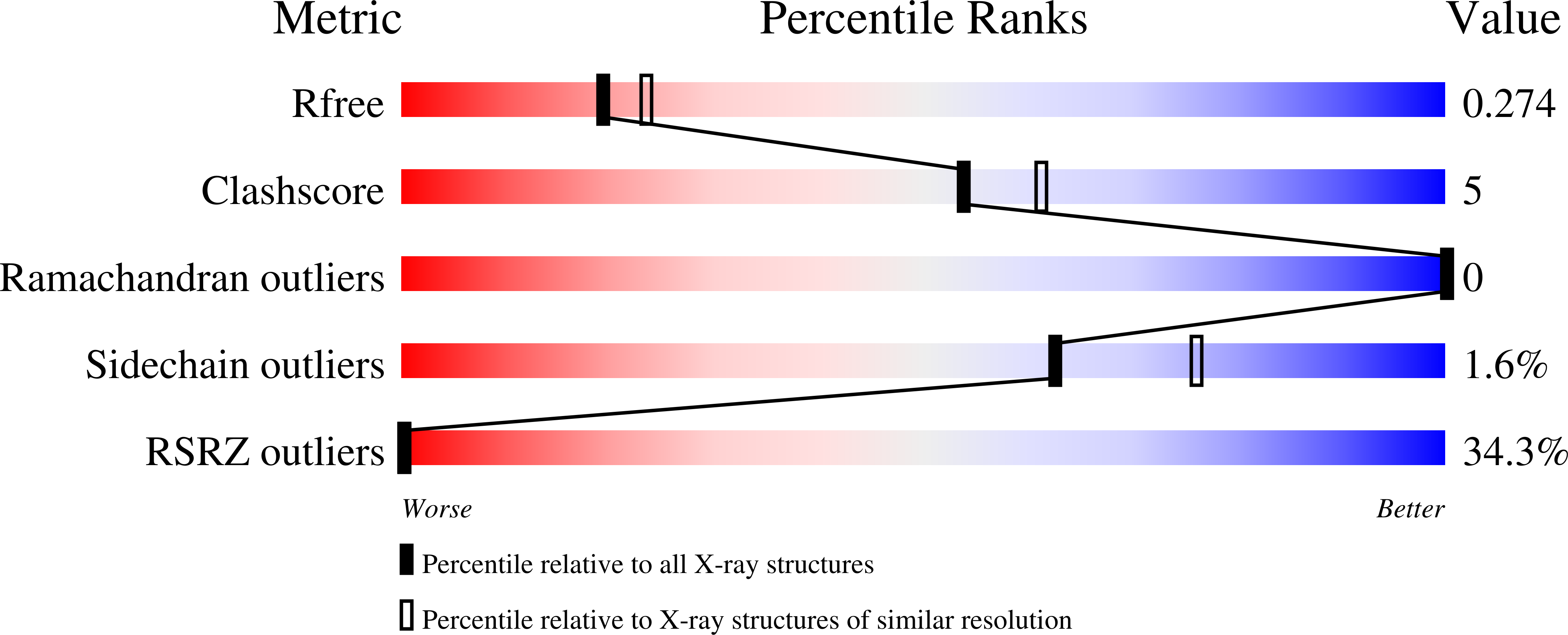
6SQC - PubMed Abstract:
Intrinsically disordered proteins (IDPs) constitute a large portion of "Dark Proteome" - difficult to characterize or yet to be discovered protein structures. Here we used conformationally constrained α-methylated amino acids to bias the conformational ensemble in the free unstructured activation domain of transcriptional coactivator ACTR. Different sites and patterns of substitutions were enabled by chemical protein synthesis and led to distinct populations of α-helices. A specific substitution pattern resulted in a substantially higher binding affinity to nuclear coactivator binding domain (NCBD) of CREB-binding protein, a natural binding partner of ACTR. The first X-ray structure of the modified ACTR domain - NCBD complex visualized a unique conformation of ACTR and confirmed that the key α-methylated amino acids are localized within α-helices in the bound state. This study demonstrates a strategy for characterization of individual conformational states of IDPs.
Organizational Affiliation:
Institut de Science et d'Ingénierie Supramoléculaires (ISIS), International Center for Frontier Research in Chemistry (icFRC), University of Strasbourg, CNRS, UMR 7006 Strasbourg France torbeev@unistra.fr.