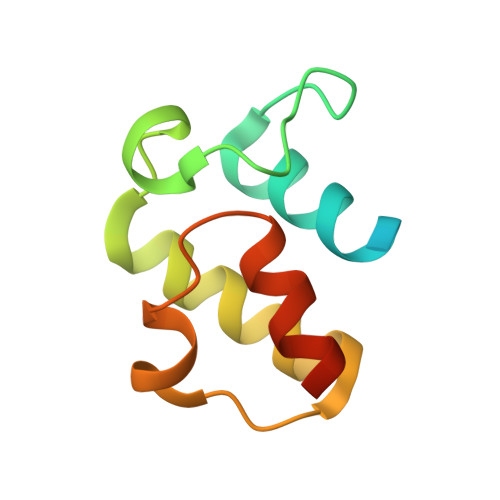
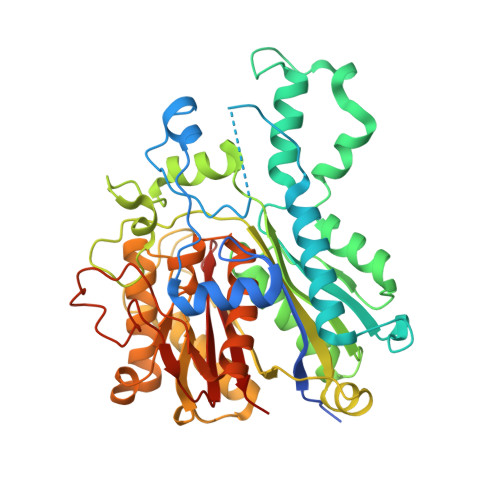
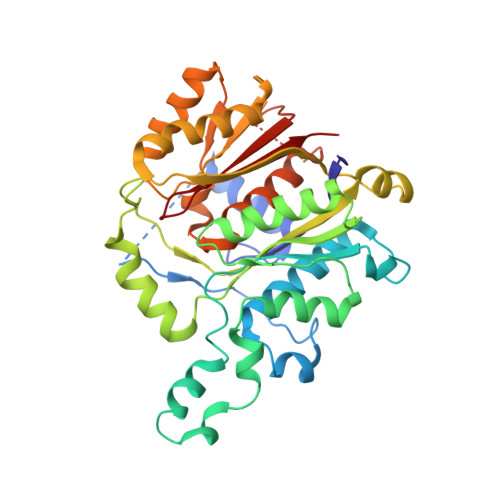
Structural snapshots of the minimal PKS system responsible for octaketide biosynthesis.
Brauer, A., Zhou, Q., Grammbitter, G.L.C., Schmalhofer, M., Ruhl, M., Kaila, V.R.I., Bode, H.B., Groll, M.(2020) Nat Chem 12: 755-763
- PubMed: 32632186
- DOI: https://doi.org/10.1038/s41557-020-0491-7
- Primary Citation of Related Structures:
6SM4, 6SM6, 6SMD, 6SMO, 6SMP - PubMed Abstract:
Type II polyketide synthases (PKSs) are multi-enzyme complexes that produce secondary metabolites of medical relevance. Chemical backbones of such polyketides are produced by minimal PKS systems that consist of a malonyl transacylase, an acyl carrier protein and an α/β heterodimeric ketosynthase. Here, we present X-ray structures of all ternary complexes that constitute the minimal PKS system for anthraquinone biosynthesis in Photorhabdus luminescens. In addition, we characterize this invariable core using molecular simulations, mutagenesis experiments and functional assays. We show that malonylation of the acyl carrier protein is accompanied by major structural rearrangements in the transacylase. Principles of an ongoing chain elongation are derived from the ternary complex with a hexaketide covalently linking the heterodimeric ketosynthase with the acyl carrier protein. Our results for the minimal PKS system provide mechanistic understanding of PKSs and a fundamental basis for engineering PKS pathways for future applications.
Organizational Affiliation:
Center for Integrated Protein Science Munich (CIPSM), Department of Chemistry, Technische Universität München, Garching, Germany.