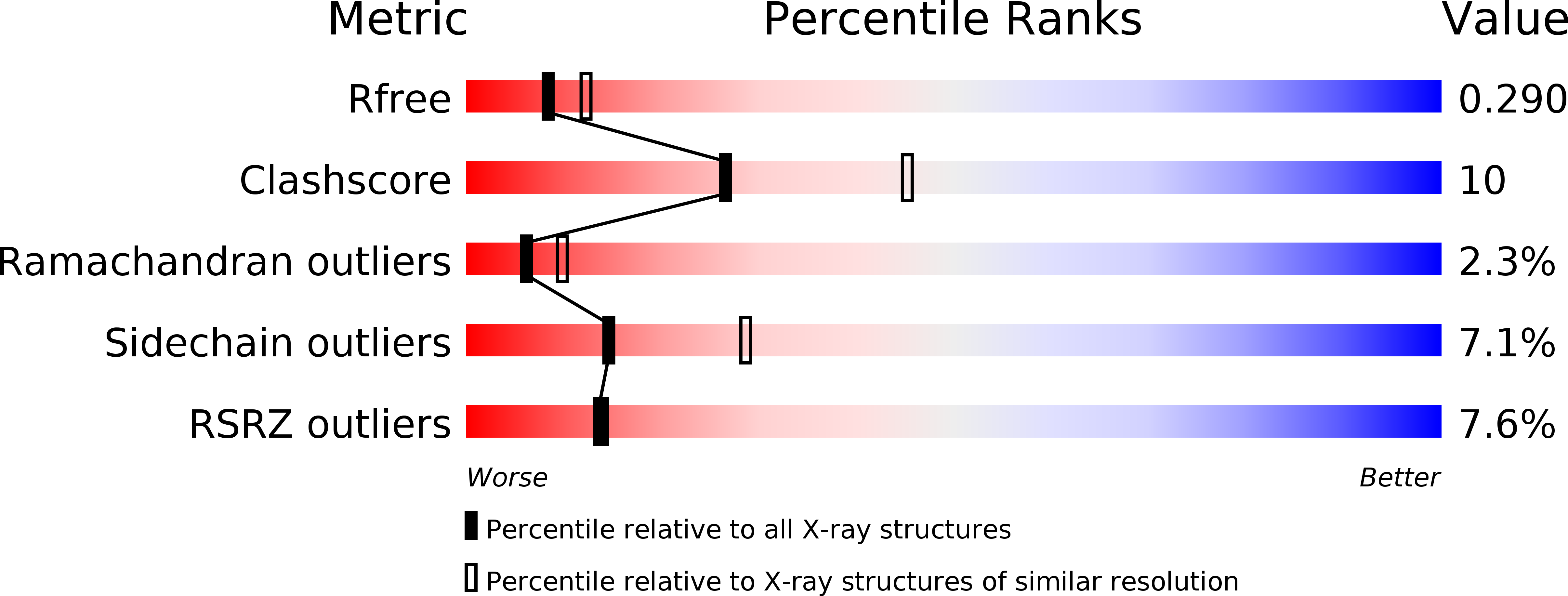
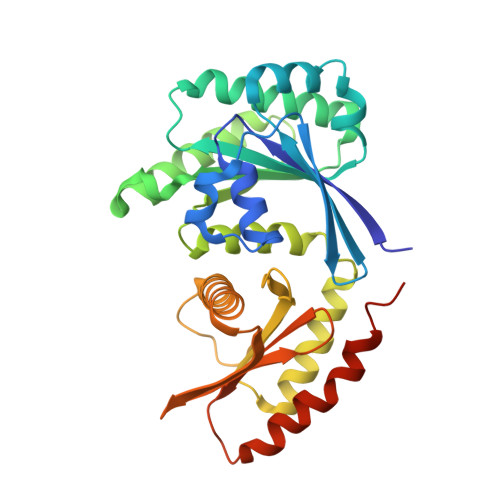
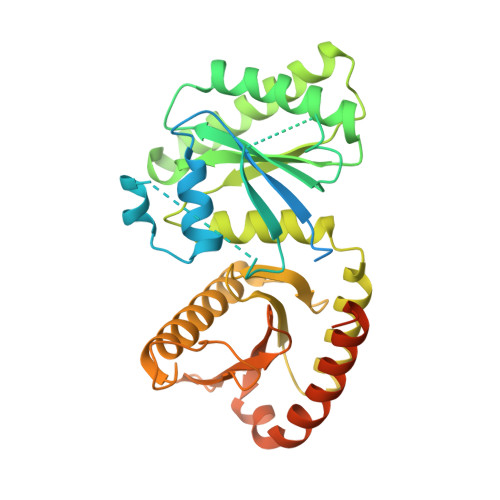
Architecture of human Rag GTPase heterodimers and their complex with mTORC1.
Anandapadamanaban, M., Masson, G.R., Perisic, O., Berndt, A., Kaufman, J., Johnson, C.M., Santhanam, B., Rogala, K.B., Sabatini, D.M., Williams, R.L.(2019) Science 366: 203-210
- PubMed: 31601764
- DOI: https://doi.org/10.1126/science.aax3939
- Primary Citation of Related Structures:
6S6A, 6S6D, 6SB0, 6SB2 - PubMed Abstract:
The Rag guanosine triphosphatases (GTPases) recruit the master kinase mTORC1 to lysosomes to regulate cell growth and proliferation in response to amino acid availability. The nucleotide state of Rag heterodimers is critical for their association with mTORC1. Our cryo-electron microscopy structure of RagA/RagC in complex with mTORC1 shows the details of RagA/RagC binding to the RAPTOR subunit of mTORC1 and explains why only the RagA GTP /RagC GDP nucleotide state binds mTORC1. Previous kinetic studies suggested that GTP binding to one Rag locks the heterodimer to prevent GTP binding to the other. Our crystal structures and dynamics of RagA/RagC show the mechanism for this locking and explain how oncogenic hotspot mutations disrupt this process. In contrast to allosteric activation by RHEB, Rag heterodimer binding does not change mTORC1 conformation and activates mTORC1 by targeting it to lysosomes.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Cambridge CB2 0QH, UK.