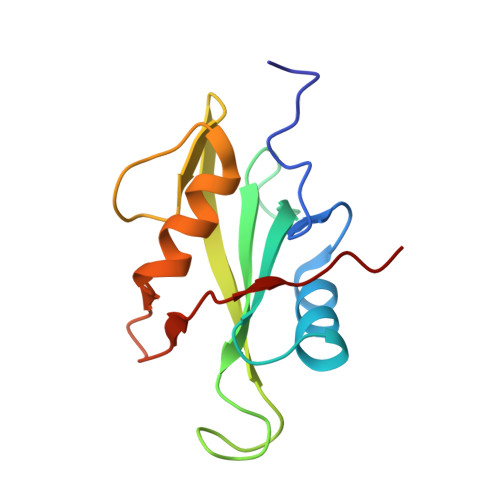
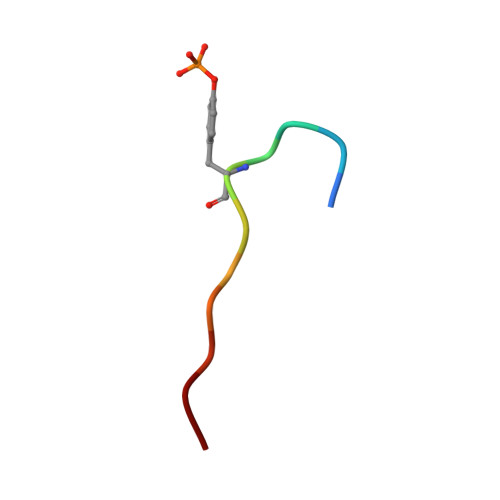
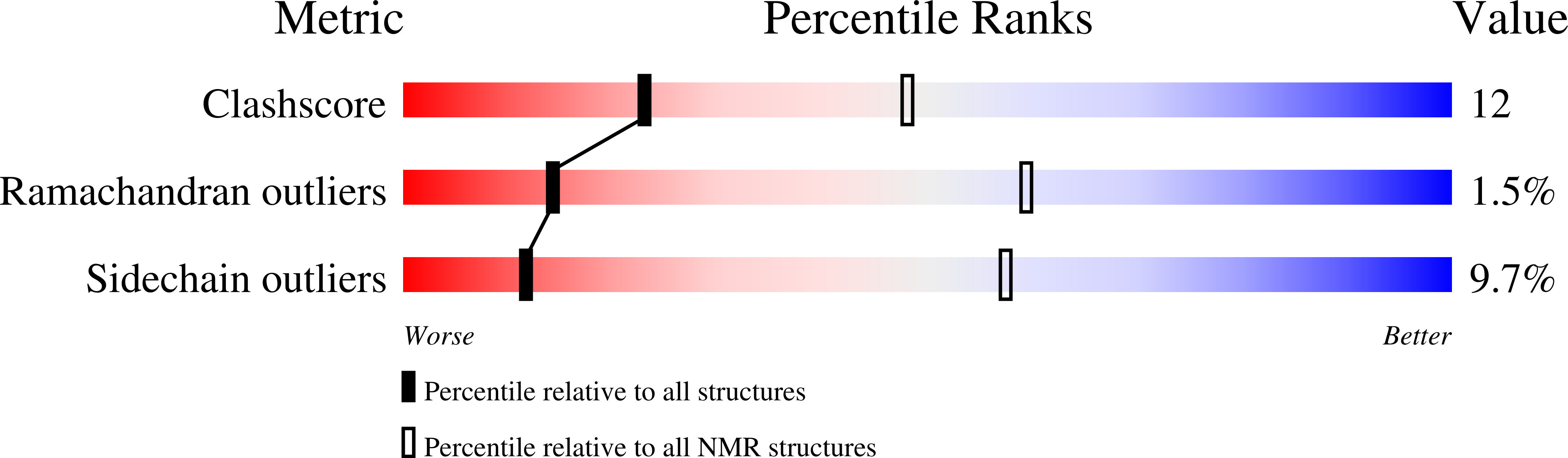Molecular mechanism of SHP2 activation by PD-1 stimulation.
Marasco, M., Berteotti, A., Weyershaeuser, J., Thorausch, N., Sikorska, J., Krausze, J., Brandt, H.J., Kirkpatrick, J., Rios, P., Schamel, W.W., Kohn, M., Carlomagno, T.(2020) Sci Adv 6: eaay4458-eaay4458
- PubMed: 32064351
- DOI: https://doi.org/10.1126/sciadv.aay4458
- Primary Citation of Related Structures:
6R5G, 6ROY, 6ROZ - PubMed Abstract:
In cancer, the programmed death-1 (PD-1) pathway suppresses T cell stimulation and mediates immune escape. Upon stimulation, PD-1 becomes phosphorylated at its immune receptor tyrosine-based inhibitory motif (ITIM) and immune receptor tyrosine-based switch motif (ITSM), which then bind the Src homology 2 (SH2) domains of SH2-containing phosphatase 2 (SHP2), initiating T cell inactivation. The SHP2-PD-1 complex structure and the exact functions of the two SH2 domains and phosphorylated motifs remain unknown. Here, we explain the structural basis and provide functional evidence for the mechanism of PD-1-mediated SHP2 activation. We demonstrate that full activation is obtained only upon phosphorylation of both ITIM and ITSM: ITSM binds C-SH2 with strong affinity, recruiting SHP2 to PD-1, while ITIM binds N-SH2, displacing it from the catalytic pocket and activating SHP2. This binding event requires the formation of a new inter-domain interface, offering opportunities for the development of novel immunotherapeutic approaches.
Organizational Affiliation:
Leibniz University Hannover, Institute of Organic Chemistry and Center for Biomolecular Drug Research, Schneiderberg 38, 30167 Hannover, Germany.