Heme binding to human CLOCK affects interactions with the E-box.
Freeman, S.L., Kwon, H., Portolano, N., Parkin, G., Venkatraman Girija, U., Basran, J., Fielding, A.J., Fairall, L., Svistunenko, D.A., Moody, P.C.E., Schwabe, J.W.R., Kyriacou, C.P., Raven, E.L.(2019) Proc Natl Acad Sci U S A 116: 19911-19916
- PubMed: 31527239
- DOI: https://doi.org/10.1073/pnas.1905216116
- Primary Citation of Related Structures:
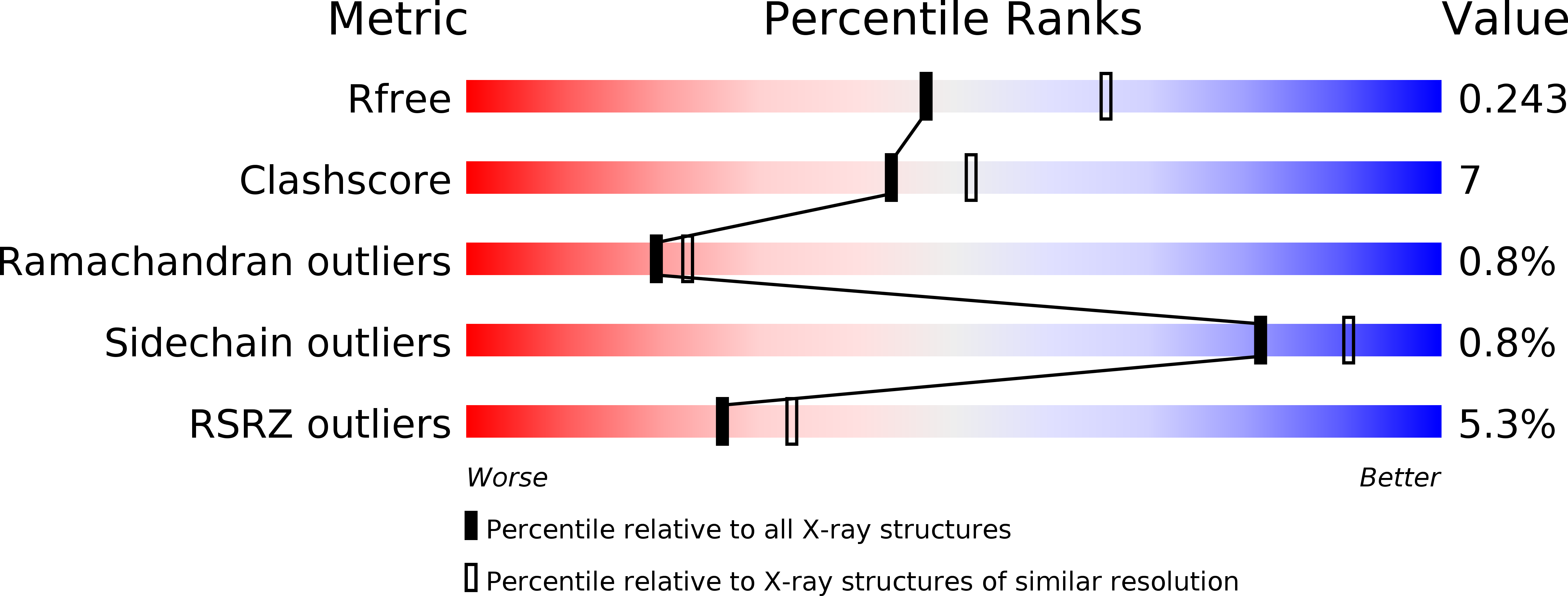
6QPJ - PubMed Abstract:
The circadian clock is an endogenous time-keeping system that is ubiquitous in animals and plants as well as some bacteria. In mammals, the clock regulates the sleep-wake cycle via 2 basic helix-loop-helix PER-ARNT-SIM (bHLH-PAS) domain proteins-CLOCK and BMAL1. There is emerging evidence to suggest that heme affects circadian control, through binding of heme to various circadian proteins, but the mechanisms of regulation are largely unknown. In this work we examine the interaction of heme with human CLOCK (hCLOCK). We present a crystal structure for the PAS-A domain of hCLOCK, and we examine heme binding to the PAS-A and PAS-B domains. UV-visible and electron paramagnetic resonance spectroscopies are consistent with a bis-histidine ligated heme species in solution in the oxidized (ferric) PAS-A protein, and by mutagenesis we identify His144 as a ligand to the heme. There is evidence for flexibility in the heme pocket, which may give rise to an additional Cys axial ligand at 20K (His/Cys coordination). Using DNA binding assays, we demonstrate that heme disrupts binding of CLOCK to its E-box DNA target. Evidence is presented for a conformationally mobile protein framework, which is linked to changes in heme ligation and which has the capacity to affect binding to the E-box. Within the hCLOCK structural framework, this would provide a mechanism for heme-dependent transcriptional regulation.
Organizational Affiliation:
School of Chemistry, University of Bristol, BS8 1TS Bristol, United Kingdom.