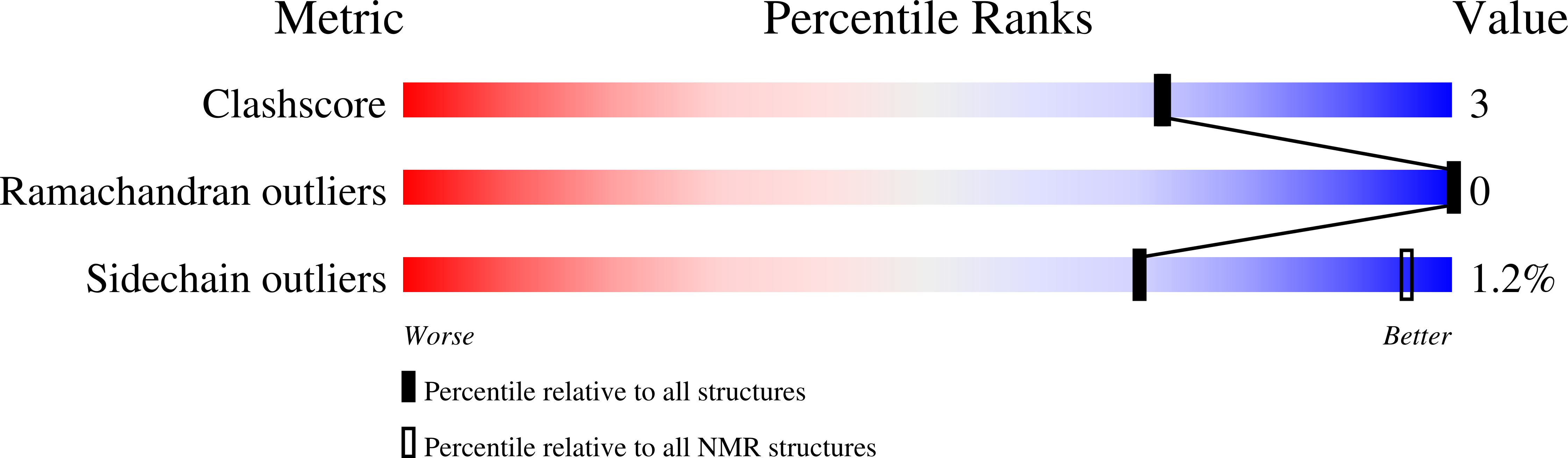A Disorder-to-Order Transition Mediates RNA Binding of the Caenorhabditis elegans Protein MEX-5.
Tavella, D., Ertekin, A., Schaal, H., Ryder, S.P., Massi, F.(2020) Biophys J 118: 2001-2014
- PubMed: 32294479
- DOI: https://doi.org/10.1016/j.bpj.2020.02.032
- Primary Citation of Related Structures:
6PMG - PubMed Abstract:
CCCH-type tandem zinc finger (TZF) domains are found in many RNA-binding proteins (RBPs) that regulate the essential processes of post-transcriptional gene expression and splicing through direct protein-RNA interactions. In Caenorhabditis elegans, RBPs control the translation, stability, or localization of maternal messenger RNAs required for patterning decisions before zygotic gene activation. MEX-5 (Muscle EXcess) is a C. elegans protein that leads a cascade of RBP localization events that is essential for axis polarization and germline differentiation after fertilization. Here, we report that at room temperature, the CCCH-type TZF domain of MEX-5 contains an unstructured zinc finger that folds upon binding of its RNA target. We have characterized the structure and dynamics of the TZF domain of MEX-5 and designed a variant MEX-5 in which both fingers are fully folded in the absence of RNA. Within the thermal range experienced by C. elegans, the population of the unfolded state of the TZF domain of MEX-5 varies. We observe that the TZF domain becomes less disordered at lower temperatures and more disordered at higher temperatures. However, in the temperature range in which C. elegans is fertile, when MEX-5 needs to be functional, only one of the two zinc fingers is folded.
Organizational Affiliation:
Department of Biochemistry and Molecular Pharmacology, University of Massachusetts Medical School, Worcester, Massachusetts.