Visualization of two architectures in class-II CAP-dependent transcription activation
Shi, W., Jiang, Y., Deng, Y., Dong, Z., Liu, B.(2020) PLoS Biol 18: e3000706
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit alpha | 329 | Escherichia coli | Mutation(s): 0 Gene Names: rpoA, Z4665, ECs4160 EC: 2.7.7.6 | 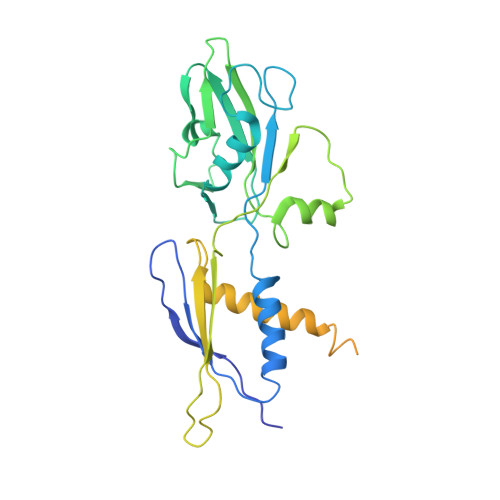 | |
UniProt | |||||
Find proteins for P0A7Z4 (Escherichia coli (strain K12)) Explore P0A7Z4 Go to UniProtKB: P0A7Z4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7Z4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit beta | 1,342 | Escherichia coli | Mutation(s): 0 Gene Names: rpoB, ECS88_4448 EC: 2.7.7.6 | 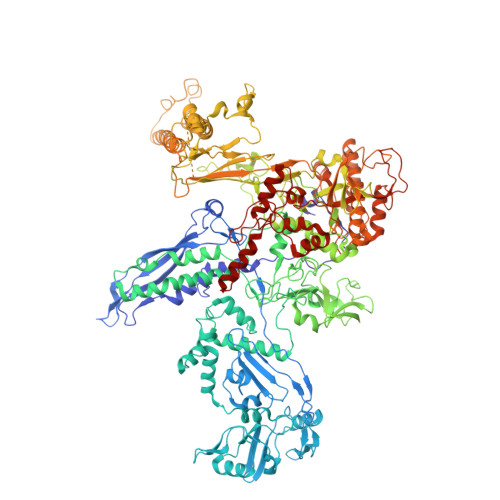 | |
UniProt | |||||
Find proteins for P0A8V2 (Escherichia coli (strain K12)) Explore P0A8V2 Go to UniProtKB: P0A8V2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A8V2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit beta' | 1,407 | Escherichia coli | Mutation(s): 0 Gene Names: rpoC, Z5561, ECs4911 EC: 2.7.7.6 | 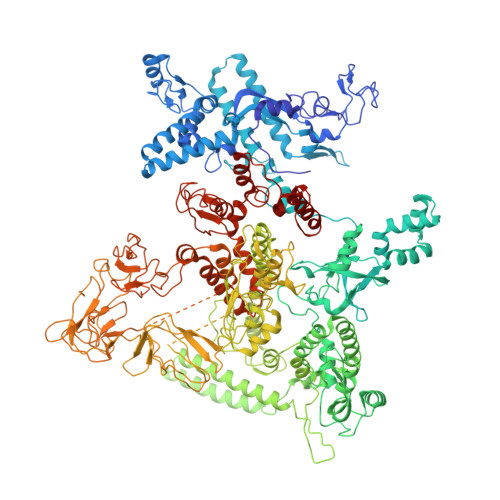 | |
UniProt | |||||
Find proteins for P0A8T7 (Escherichia coli (strain K12)) Explore P0A8T7 Go to UniProtKB: P0A8T7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A8T7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA-directed RNA polymerase subunit omega | 91 | Escherichia coli | Mutation(s): 0 Gene Names: rpoZ, ECS88_4064 EC: 2.7.7.6 | 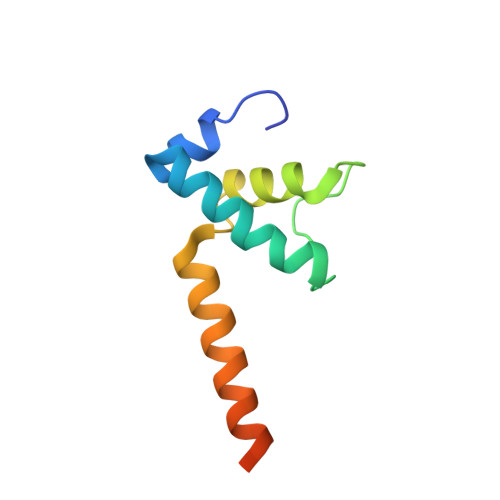 | |
UniProt | |||||
Find proteins for P0A800 (Escherichia coli (strain K12)) Explore P0A800 Go to UniProtKB: P0A800 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A800 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNA polymerase sigma factor RpoD | 628 | Escherichia coli | Mutation(s): 0 Gene Names: rpoD, alt, b3067, JW3039 | 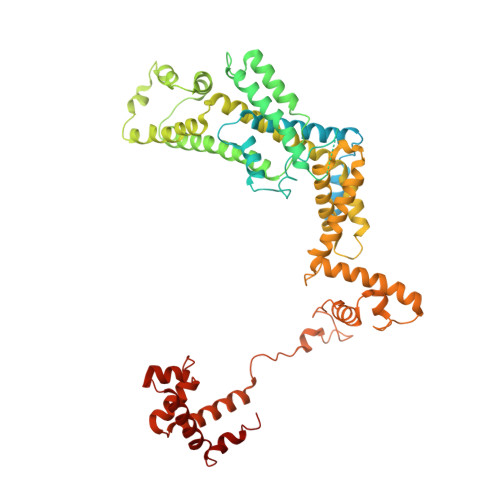 | |
UniProt | |||||
Find proteins for P00579 (Escherichia coli (strain K12)) Explore P00579 Go to UniProtKB: P00579 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00579 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| cAMP-activated global transcriptional regulator CRP | 210 | Escherichia coli | Mutation(s): 0 Gene Names: crp, Z4718, ECs4208 | 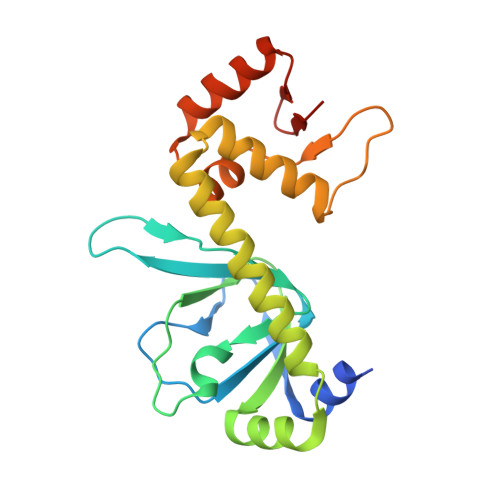 | |
UniProt | |||||
Find proteins for P0ACJ8 (Escherichia coli (strain K12)) Explore P0ACJ8 Go to UniProtKB: P0ACJ8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ACJ8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| SYNTHETIC NONTEMPLATE STRAND DNA (78-MER) | I [auth 1] | 78 | Escherichia coli | 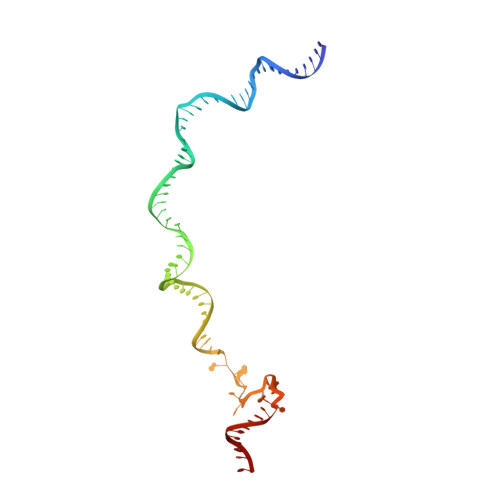 | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| SYNTHETIC TEMPLATE STRAND DNA (78-MER) | J [auth 2] | 78 | Escherichia coli | 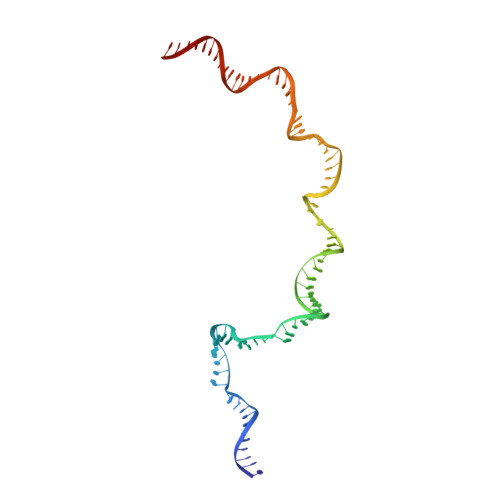 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CMP (Subject of Investigation/LOI) Query on CMP | N [auth G], O [auth H] | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE C10 H12 N5 O6 P IVOMOUWHDPKRLL-KQYNXXCUSA-N |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | K [auth D], L [auth D] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | M [auth D] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cisTEM |