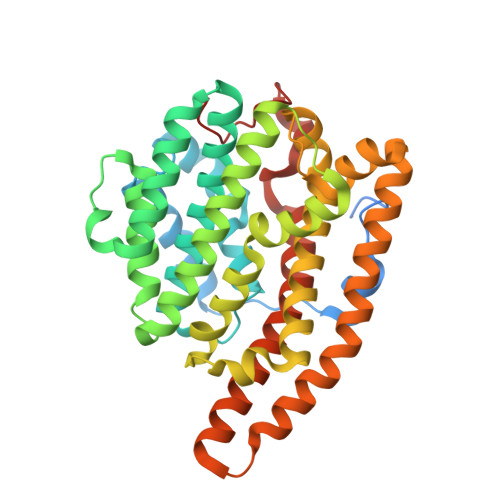
Crystal structure of F95Q epi-isozizaene synthase, an engineered sesquiterpene cyclase that generates biofuel precursors beta- and gamma-curcumene.
Blank, P.N., Barrow, G.H., Christianson, D.W.(2019) J Struct Biol 207: 218-224
- PubMed: 31152775
- DOI: https://doi.org/10.1016/j.jsb.2019.05.011
- Primary Citation of Related Structures:
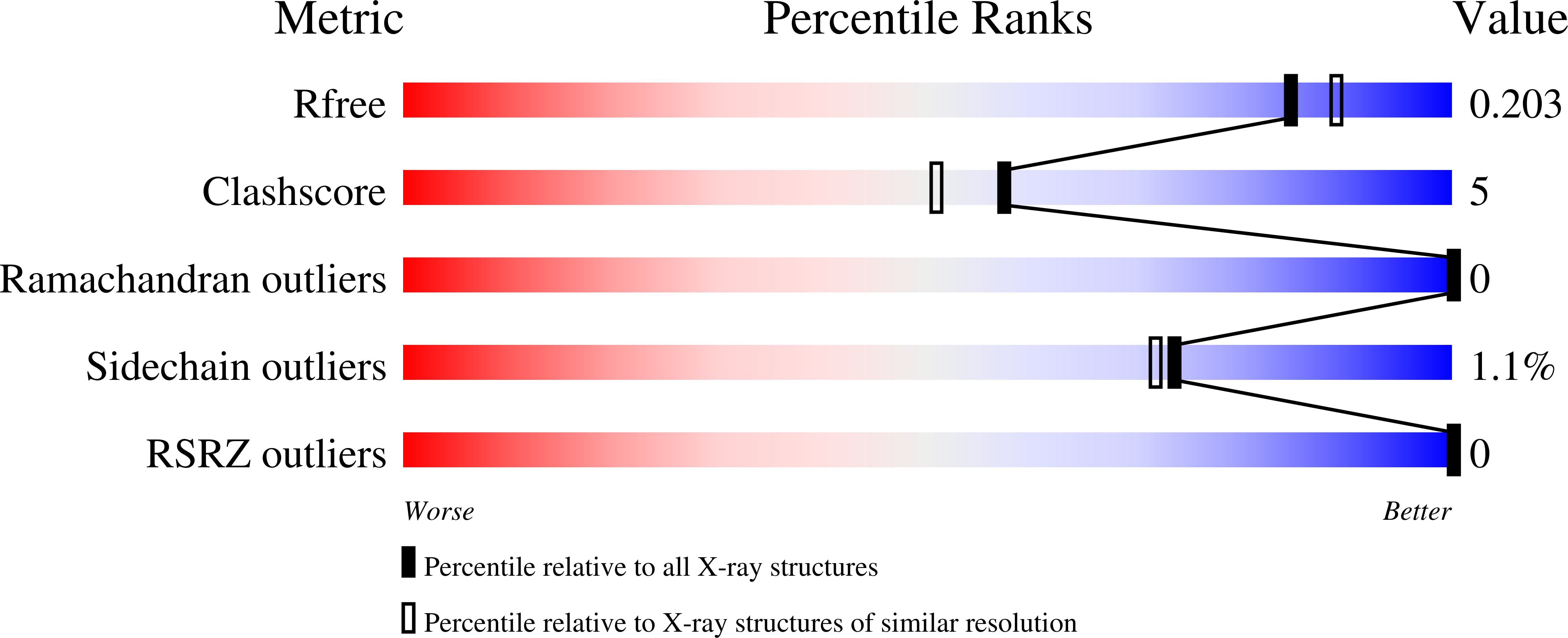
6OFV - PubMed Abstract:
The saturated hydrocarbon bisabolane is a diesel fuel substitute that can be derived from sesquiterpene precursors bisabolene or curcumene. These sesquiterpenes are generated from farnesyl diphosphate in reactions catalyzed by eponymous terpenoid cyclases, but they can also be generated by engineered terpenoid cyclases in which cyclization cascades have been reprogrammed by mutagenesis. Here, we describe the X-ray crystal structure determination of F95Q epi-isozizaene synthase (EIZS), in which the new activity of curcumene biosynthesis has been introduced and the native activity of epi-isozizaene biosynthesis has been suppressed. F95Q EIZS generates β- and γ-curcumene regioisomers with greater than 50% yield. Structural analysis of the closed active site conformation, stabilized by the binding of 3 Mg 2+ ions, inorganic pyrophosphate, and the benzyltriethylammonium cation, reveals a product-like active site contour that serves as the cyclization template. Remolding the active site contour to resemble curcumene instead of epi-isozizaene is the principal determinant of the reprogrammed cyclization cascade. Intriguingly, an ordered water molecule comprises part of the active site contour. This water molecule may also serve as a final proton acceptor, along with inorganic pyrophosphate, in the generation of curcumene regioisomers; it may also contribute to the formation of sesquiterpene alcohols identified as minor side products. Thus, the substitution of polar side chains for nonpolar side chains in terpenoid cyclase active sites can result in the stabilization of bound water molecules that, in turn, can serve template functions in isoprenoid cyclization reactions.
Organizational Affiliation:
Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, 231 South 34(th) Street, Philadelphia, PA 19104-6323, United States.