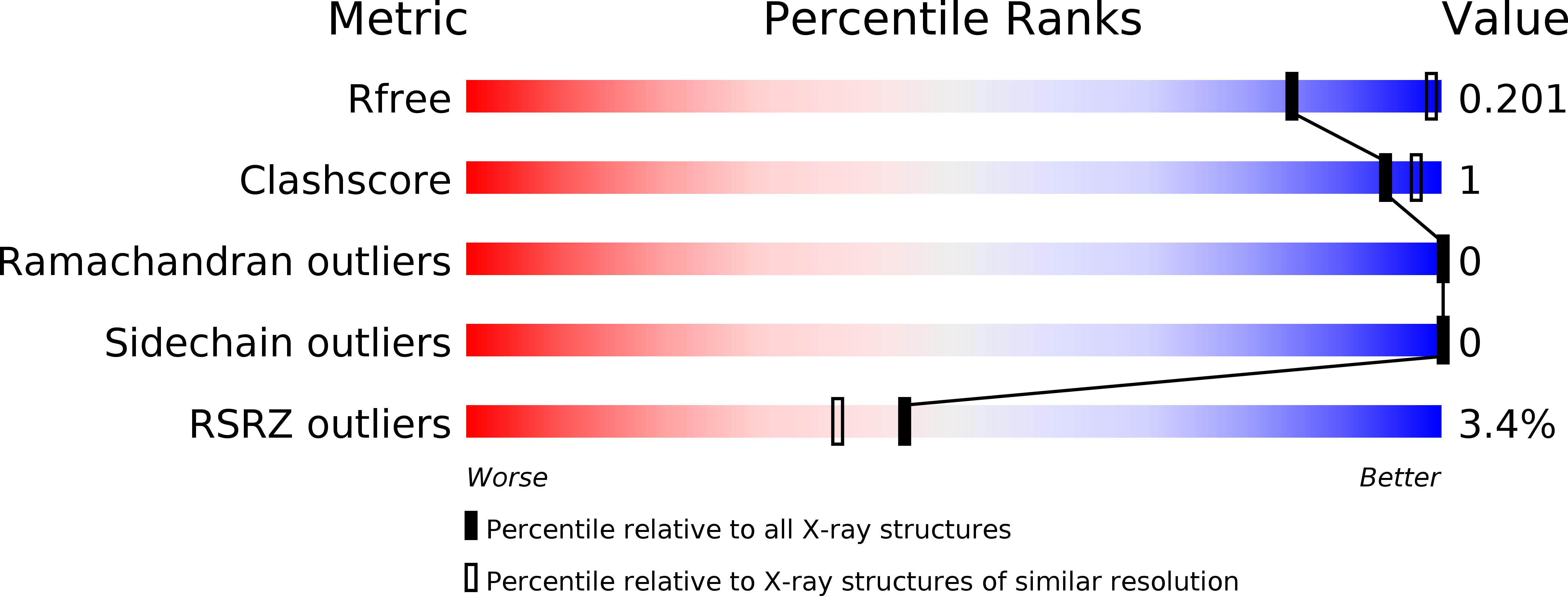
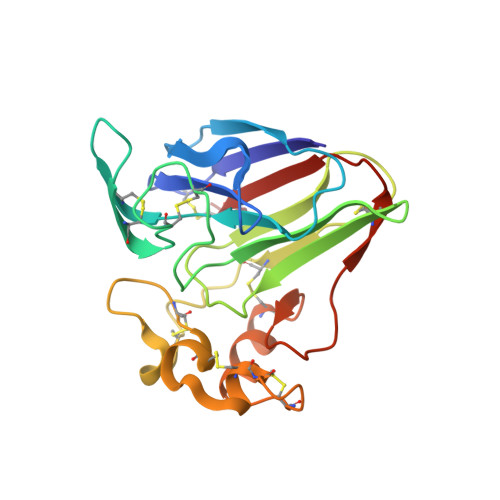
Synchrotron microcrystal native-SAD phasing at a low energy.
Guo, G., Zhu, P., Fuchs, M.R., Shi, W., Andi, B., Gao, Y., Hendrickson, W.A., McSweeney, S., Liu, Q.(2019) IUCrJ 6: 532-542
- PubMed: 31316798
- DOI: https://doi.org/10.1107/S2052252519004536
- Primary Citation of Related Structures:
6O8A - PubMed Abstract:
De novo structural evaluation of native biomolecules from single-wavelength anomalous diffraction (SAD) is a challenge because of the weakness of the anomalous scattering. The anomalous scattering from relevant native elements - primarily sulfur in proteins and phospho-rus in nucleic acids - increases as the X-ray energy decreases toward their K -edge transitions. Thus, measurements at a lowered X-ray energy are promising for making native SAD routine and robust. For microcrystals with sizes less than 10 µm, native-SAD phasing at synchrotron microdiffraction beamlines is even more challenging because of difficulties in sample manipulation, diffraction data collection and data analysis. Native-SAD analysis from microcrystals by using X-ray free-electron lasers has been demonstrated but has required use of thousands of thousands of microcrystals to achieve the necessary accuracy. Here it is shown that by exploitation of anomalous microdiffraction signals obtained at 5 keV, by the use of polyimide wellmounts, and by an iterative crystal and frame-rejection method, microcrystal native-SAD phasing is possible from as few as about 1 200 crystals. Our results show the utility of low-energy native-SAD phasing with microcrystals at synchrotron microdiffraction beamlines.
Organizational Affiliation:
Biology Department, Brookhaven National Laboratory, Upton, NY 11973, USA.