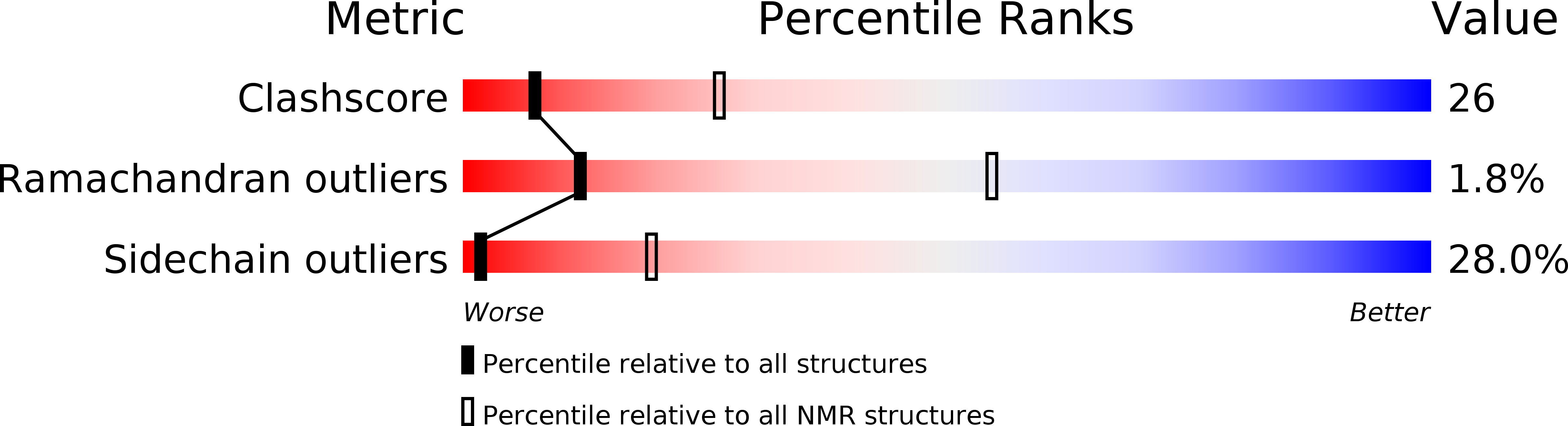Low cationicity is important for systemic in vivo efficacy of database-derived peptides against drug-resistant Gram-positive pathogens.
Mishra, B., Lakshmaiah Narayana, J., Lushnikova, T., Wang, X., Wang, G.(2019) Proc Natl Acad Sci U S A 116: 13517-13522
- PubMed: 31209048
- DOI: https://doi.org/10.1073/pnas.1821410116
- Primary Citation of Related Structures:
6MK8 - PubMed Abstract:
As bacterial resistance to traditional antibiotics continues to emerge, new alternatives are urgently needed. Antimicrobial peptides (AMPs) are important candidates. However, how AMPs are designed with in vivo efficacy is poorly understood. Our study was designed to understand structural moieties of cationic peptides that would lead to their successful use as antibacterial agents. In contrast to the common perception, serum binding and peptide stability were not the major reasons for in vivo failure in our studies. Rather, our systematic study of a series of peptides with varying lysines revealed the significance of low cationicity for systemic in vivo efficacy against Gram-positive pathogens. We propose that peptides with biased amino acid compositions are not favored to associate with multiple host factors and are more likely to show in vivo efficacy. Thus, our results uncover a useful design strategy for developing potent peptides against multidrug-resistant pathogens.
Organizational Affiliation:
Department of Pathology and Microbiology, College of Medicine, University of Nebraska Medical Center, Omaha, NE 68198-5900.