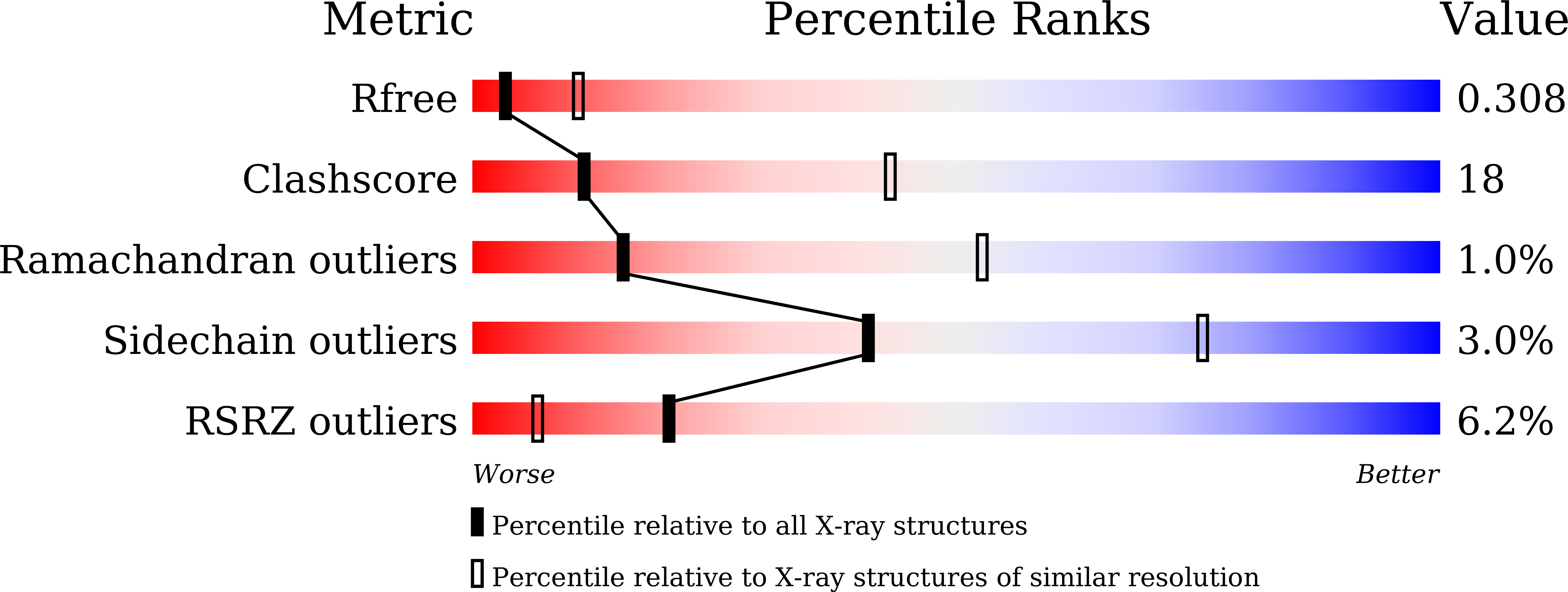
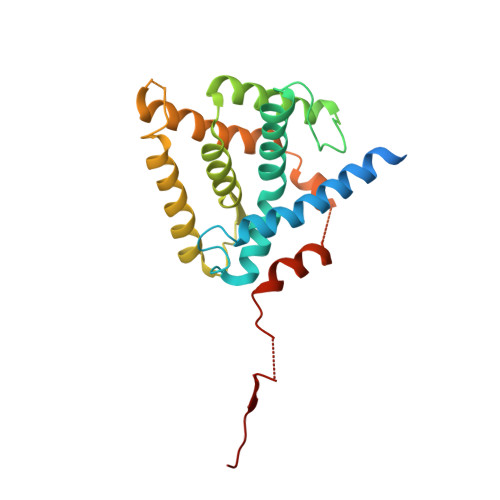
Crystal structure of the mineralocorticoid receptor ligand-binding domain in complex with a potent and selective nonsteroidal blocker, esaxerenone (CS-3150).
Takahashi, M., Ubukata, O., Homma, T., Asoh, Y., Honzumi, M., Hayashi, N., Saito, K., Tsuruoka, H., Aoki, K., Hanzawa, H.(2020) FEBS Lett 594: 1615-1623
- PubMed: 31991486
- DOI: https://doi.org/10.1002/1873-3468.13746
- Primary Citation of Related Structures:
6L88 - PubMed Abstract:
Activation of the mineralocorticoid receptor (MR) has long been considered a risk factor for cardiovascular diseases. It has been reported that the novel MR blocker esaxerenone shows high potency and selectivity for MR in vitro as well as great antihypertensive and renoprotective effects in salt-sensitive hypertensive rats. Here, we determined the cocrystal structure of the MR ligand-binding domain (MR-LBD) with esaxerenone and found that esaxerenone binds to MR-LBD in a unique manner with large side-chain rearrangements, distinct from those of previously published MR antagonists. This structure also displays an antagonist form that has not been observed for MR previously. Such a unique binding mode of esaxerenone provides great insight into the novelty, potency, and selectivity of this novel antihypertensive drug.
Organizational Affiliation:
Structure-Based Drug Design Group, Organic Synthesis Department, Daiichi Sankyo RD Novare Co., Ltd., Tokyo, Japan.