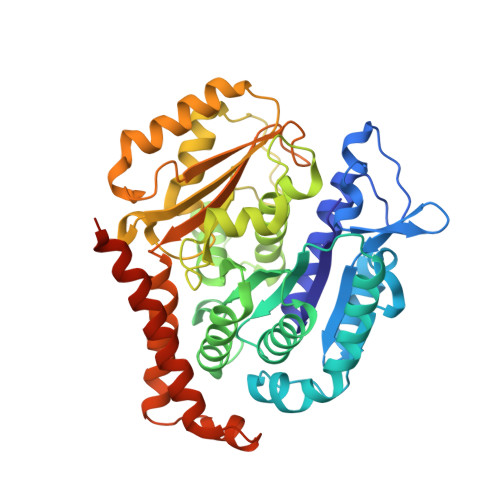
Unraveling the molecular mechanism of BNC105, a phase II clinical trial vascular disrupting agent, provides insights into drug design.
Wang, T., Wu, C., Wang, C., Zhang, G., Arnst, K.E., Yao, Y., Zhang, Z., Wang, Y., Pu, D., Li, W.(2020) Biochem Biophys Res Commun
- PubMed: 32085900
- DOI: https://doi.org/10.1016/j.bbrc.2019.12.083
- Primary Citation of Related Structures:
6KPP - PubMed Abstract:
Microtubules are made up of tubulin protein and play a very important part in numerous cellular events of eukaryotic cells, which is why they are seen as attractive targets for tumor chemotherapy. BNC105, a known vascular targeting agent, has entered in phase II clinical trials. It has previously been confirmed that BNC105 is an effective microtubule targeting agent for various cancers. BNC105 exhibits selectivity for tumor cells, elicits vascular disrupting effects, and inhibits tumor growth. However, the molecular mechanism of BNC105 is still elusive. Herein, the crystal structure of BNC105 in complex with tubulin protein is revealed, demonstrating the its interaction with the colchicine binding site. In order to thoroughly evaluate its molecular mechanism from a structural-activity-relationship standpoint, the binding mode of tubulin to BNC-105 is compared with colchicine, CA-4 and other BNC-105 derivatives. Our study not only confirms the detailed interactions of the BNC105-tubulin complex, but also offer substantial structural foundation for the design and development of novel benzo[b]furan derivatives as microtubule targeting agents.
Organizational Affiliation:
Department of Respiratory and Critical Care Medicine, West China Hospital of Sichuan University, Chengdu, Sichuan, 610041, PR China; Department of Respiratory and Critical Care Medicine, Affiliated Hospital of Zunyi Medical University, Zunyi, Guizhou, PR China.