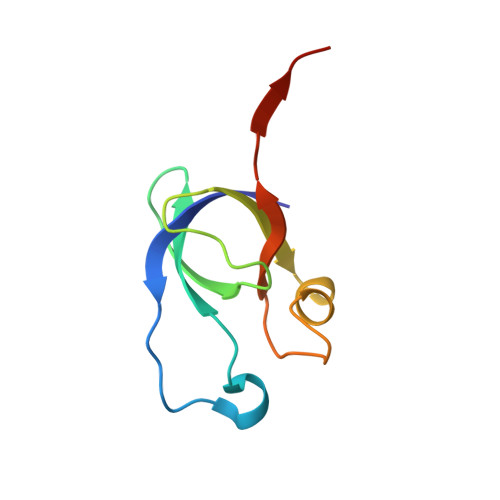
Crystal structure of pentameric shell protein CsoS4B of Halothiobacillus neapolitanus alpha-carboxysome.
Zhao, Y.Y., Jiang, Y.L., Chen, Y., Zhou, C.Z., Li, Q.(2019) Biochem Biophys Res Commun 515: 510-515
- PubMed: 31171360
- DOI: https://doi.org/10.1016/j.bbrc.2019.05.047
- Primary Citation of Related Structures:
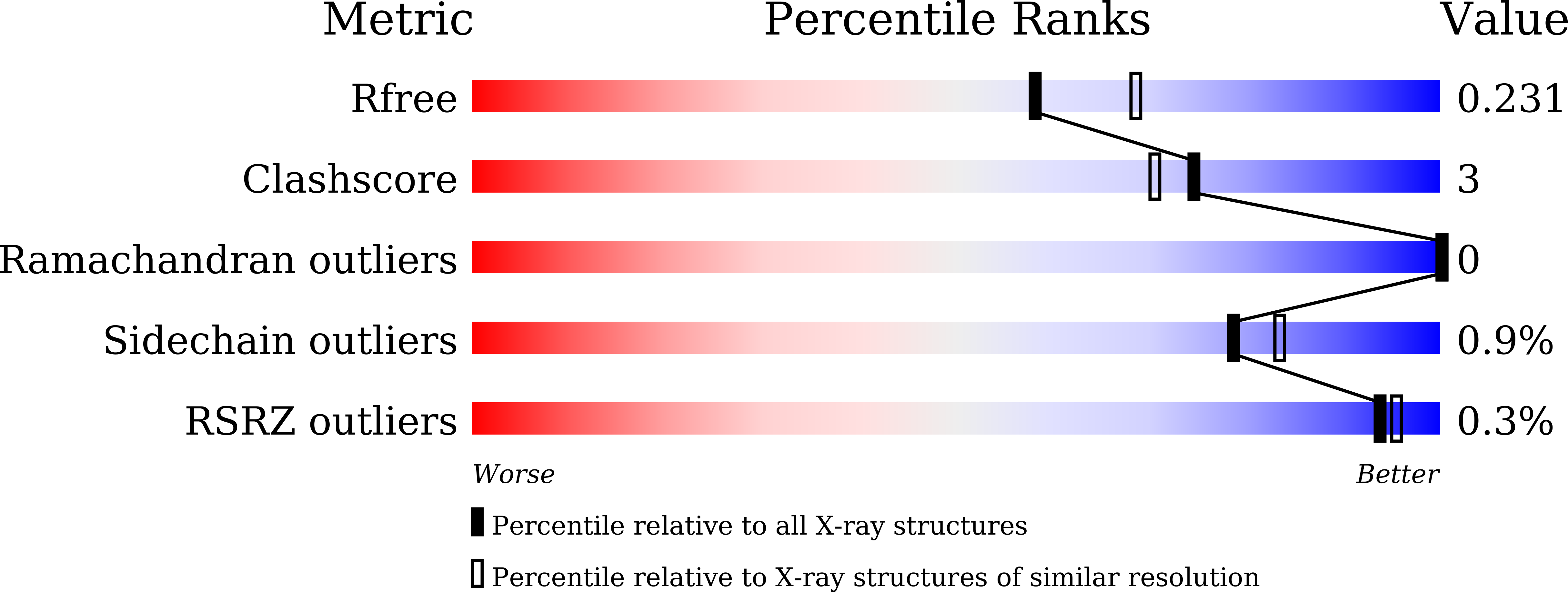
6JY5 - PubMed Abstract:
Carboxysome, encapsulating an enzymatic core within an icosahedral-shaped semipermeable protein shell, could enhance CO 2 fixation under low CO 2 conditions in the environment. The shell of Halothiobacillus neapolitanus α-carboxysome possesses two 38% sequence-identical pentameric proteins, namely CsoS4A and CsoS4B. However, the functions of two paralogous pentameric proteins in α-carboxysome assembly remain unknown. Here we report the crystal structure of CsoS4B at 2.15 Å resolution. It displays as a stable pentamer, each subunit of which consists of a β-barrel core domain, in addition to an insertion of helix α1 that forms the central pore. Structural comparisons and multiple-sequence alignment strongly indicate that CsoS4A and CsoS4B differ from each other in interacting with various components of α-carboxysome, despite they share a similar overall structure. These findings provide the structural basis for further investigations on the self-assembly process of carboxysome.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui, 230026, China.