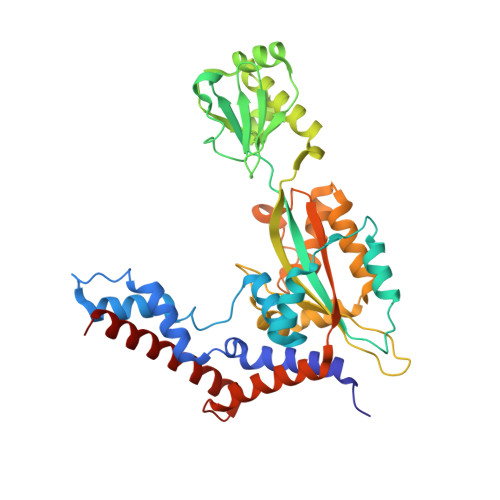
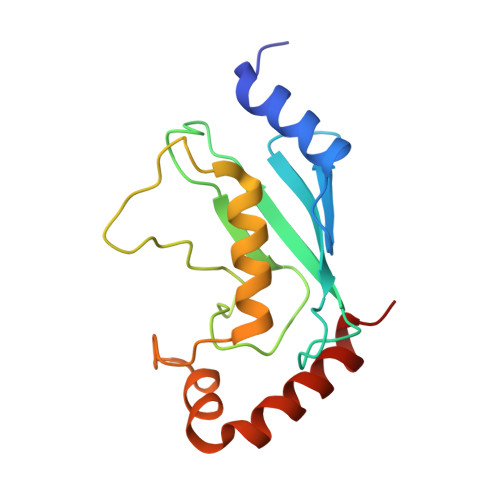
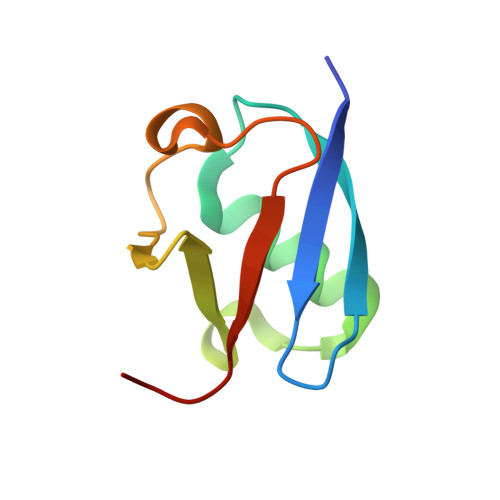
Legionella pneumophila regulates the activity of UBE2N by deamidase-mediated deubiquitination.
Gan, N., Guan, H., Huang, Y., Yu, T., Fu, J., Nakayasu, E.S., Puvar, K., Das, C., Wang, D., Ouyang, S., Luo, Z.Q.(2020) EMBO J 39: e102806-e102806
- PubMed: 31825121
- DOI: https://doi.org/10.15252/embj.2019102806
- Primary Citation of Related Structures:
6JKY, 6K11 - PubMed Abstract:
The Legionella pneumophila effector MavC induces ubiquitination of the E2 ubiquitin-conjugating enzyme UBE2N by transglutamination, thereby abolishing its function in the synthesis of K 63 -type polyubiquitin chains. The inhibition of UBE2N activity creates a conundrum because this E2 enzyme is important in multiple signaling pathways, including some that are important for intracellular L. pneumophila replication. Here, we show that prolonged inhibition of UBE2N activity by MavC restricts intracellular bacterial replication and that the activity of UBE2N is restored by MvcA, an ortholog of MavC (50% identity) with ubiquitin deamidase activity. MvcA functions to deubiquitinate UBE2N-Ub using the same catalytic triad required for its deamidase activity. Structural analysis of the MvcA-UBE2N-Ub complex reveals a crucial role of the insertion domain in MvcA in substrate recognition. Our study establishes a deubiquitination mechanism catalyzed by a deamidase, which, together with MavC, imposes temporal regulation of the activity of UBE2N during L. pneumophila infection.
Organizational Affiliation:
Purdue Institute for Inflammation, Immunology and Infectious Disease and Department of Biological Sciences, Purdue University, West Lafayette, IN, USA.