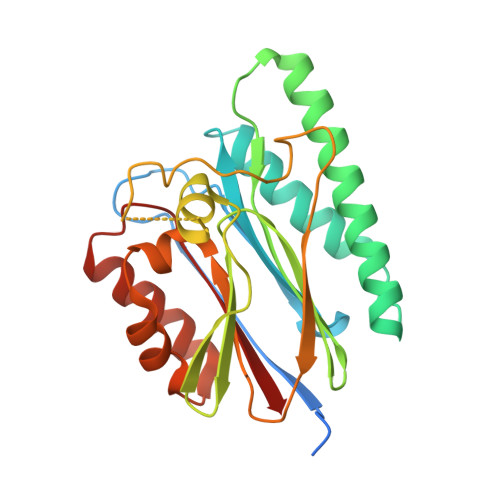
Crystal structure of PppA from Pseudomonas aeruginosa, a key regulatory component of type VI secretion systems.
Wu, Y., Gong, J., Liu, S., Li, D., Wu, Y., Zhang, X., Ren, Y., Xu, S., Sun, J., Wang, T., Lin, Q., Liu, L.(2019) Biochem Biophys Res Commun 516: 196-201
- PubMed: 31208722
- DOI: https://doi.org/10.1016/j.bbrc.2019.06.020
- Primary Citation of Related Structures:
6JKV - PubMed Abstract:
The Type VI secretion system (T6SS) is a membrane protein complex related to inter-bacterial competitions and host-pathogen interactions in Pseudomonas aeruginosa. The T6SS is regulated by a great variety of regulatory mechanisms at multiple levels, including post-translational modification with threonine phosphorylation mediated by Ser/Thr protein kinase PpkA and phosphatase PppA. The T6SS is activated by PpkA via Thr phosphorylation of Fha, and PppA can antagonize PpkA. PppA is a PP2C-family protein phosphatase and plays a key role in the disassembly and reassembly of T6SS organelles. Herein, we report the first crystal structure of PppA from Pseudomonas aeruginosa, which was determined at a resolution of 2.10 Å. The overall structure consists of a bacteria PPM structural core and a flexible flap subdomain. PppA harbors a catalytic pocket containing two manganese ions which correspond to the canonical dinuclear metal center of Ser/Thr protein phosphatases including the bacterial PPM phosphatases and human PP2C. The flexibility and the diversity of the sequence of flap subdomain across the homologues might provide clues for substrates specific recognition of phosphatases.
Organizational Affiliation:
School of Chemical Biology and Biotechnology, Peking University Shenzhen Graduate School, Shenzhen, 518055, China; Department of Biology, Southern University of Science and Technology, Shenzhen, 518055, China.