Formyl-Met-Ala-Ser bound crystal structure of class II peptide deformylase from methicillin resistant Staphylococcus aureus
Lee, I.H., Ho, T.H., Kang, L.W.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peptide deformylase | 183 | Staphylococcus aureus | Mutation(s): 0 Gene Names: def, def1, pdf1 EC: 3.5.1.88 | 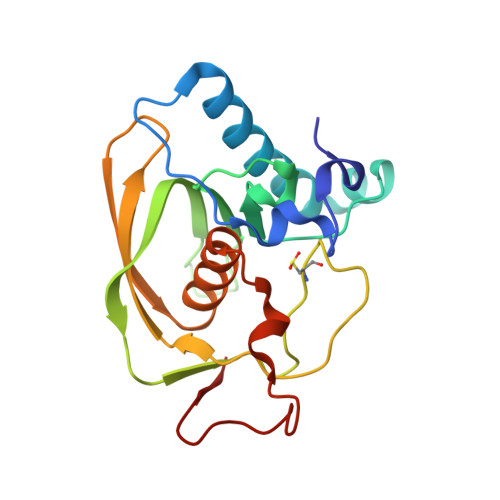 | |
UniProt | |||||
Find proteins for P68826 (Staphylococcus aureus) Explore P68826 Go to UniProtKB: P68826 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P68826 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| FME-ALA-SER | 3 | Arabidopsis thaliana | Mutation(s): 0 | 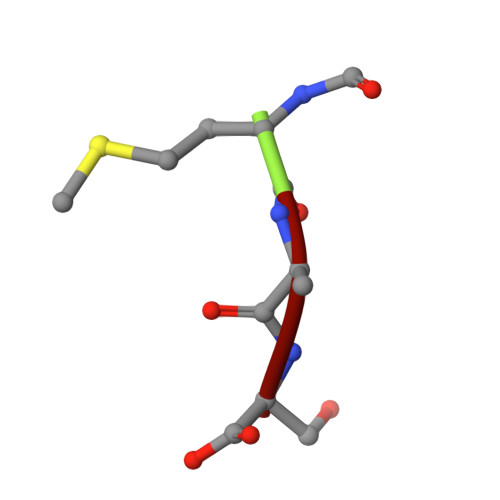 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MG Query on MG | C [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Modified Residues 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSD Query on CSD | A | L-PEPTIDE LINKING | C3 H7 N O4 S |  | CYS |
| FME Query on FME | B | L-PEPTIDE LINKING | C6 H11 N O3 S |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 94.733 | α = 90 |
| b = 120.831 | β = 90 |
| c = 47.393 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data scaling |
| REFMAC | refinement |
| HKL-2000 | data collection |
| HKL-2000 | data reduction |
| MOLREP | phasing |