High resolution crystal structure of human FLRT3 LRR domain in complex with mouse CIRL3 Olfactomedin like domain
Liu, H., Li, Z., Xu, F.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Adhesion G protein-coupled receptor L3 | 276 | Mus musculus | Mutation(s): 0 Gene Names: Adgrl3, Kiaa0768, Lec3, Lphn3 | 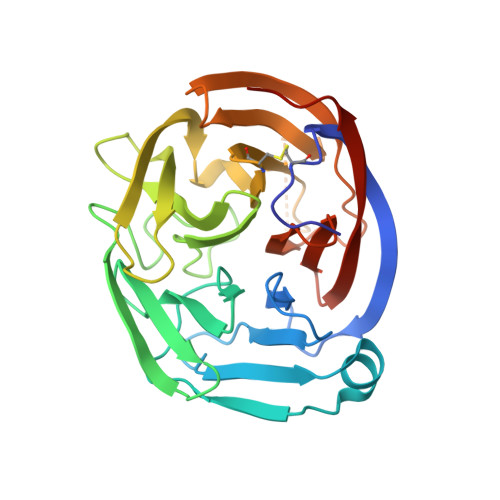 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q80TS3 (Mus musculus) Explore Q80TS3 Go to UniProtKB: Q80TS3 | |||||
IMPC: MGI:2441950 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q80TS3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Leucine-rich repeat transmembrane protein FLRT3 | 347 | Homo sapiens | Mutation(s): 0 Gene Names: FLRT3, KIAA1469, UNQ856/PRO1865 | 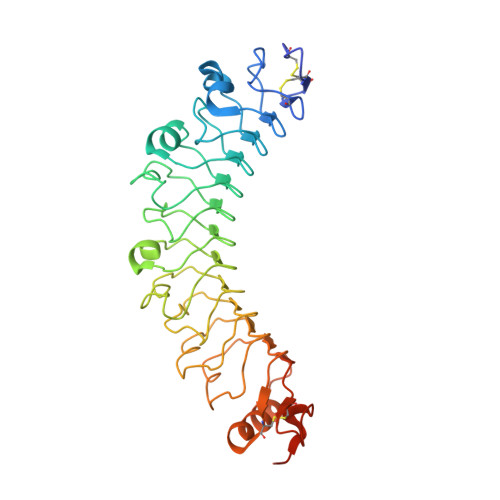 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9NZU0 (Homo sapiens) Explore Q9NZU0 Go to UniProtKB: Q9NZU0 | |||||
PHAROS: Q9NZU0 GTEx: ENSG00000125848 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9NZU0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | L [auth B], M [auth B] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| PO4 Query on PO4 | C [auth A], D [auth A] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| EDO Query on EDO | G [auth A], H [auth A], I [auth A], J [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| NA Query on NA | E [auth A], F [auth A], K [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 121.135 | α = 90 |
| b = 121.135 | β = 90 |
| c = 83.413 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data scaling |
| PHASER | phasing |