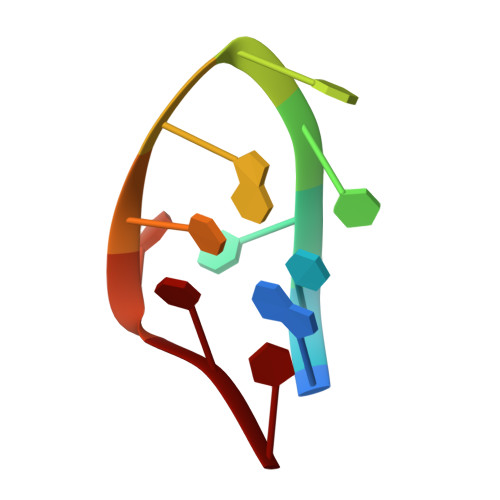
Minidumbbell structures formed by ATTCT pentanucleotide repeats in spinocerebellar ataxia type 10.
Guo, P., Lam, S.L.(2020) Nucleic Acids Res 48: 7557-7568
- PubMed: 32520333
- DOI: https://doi.org/10.1093/nar/gkaa495
- Primary Citation of Related Structures:
6IY5 - PubMed Abstract:
Spinocerebellar ataxia type 10 (SCA10) is a progressive genetic disorder caused by ATTCT pentanucleotide repeat expansions in intron 9 of the ATXN10 gene. ATTCT repeats have been reported to form unwound secondary structures which are likely linked to large-scale repeat expansions. In this study, we performed high-resolution nuclear magnetic resonance spectroscopic investigations on DNA sequences containing two to five ATTCT repeats. Strikingly, we found the first two repeats of all these sequences well folded into highly compact minidumbbell (MDB) structures. The 3D solution structure of the sequence containing two ATTCT repeats was successfully determined, revealing the MDB comprises a regular TTCTA and a quasi TTCT/A pentaloops with extensive stabilizing loop-loop interactions. We further carried out in vitro primer extension assays to examine if the MDB formed in the primer could escape from the proofreading function of DNA polymerase. Results showed that when the MDB was formed at 5-bp or farther away from the priming site, it was able to escape from the proofreading by Klenow fragment of DNA polymerase I and thus retained in the primer. The intriguing structural findings bring about new insights into the origin of genetic instability in SCA10.
Organizational Affiliation:
School of Biology and Biological Engineering, South China University of Technology, Guangzhou, Guangdong 510006, China.