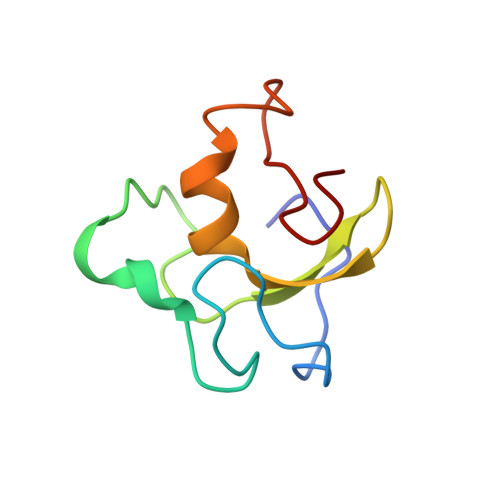
Zinc finger domain of the human DTX protein adopts a unique RING fold.
Miyamoto, K., Fujiwara, Y., Saito, K.(2019) Protein Sci 28: 1151-1156
- PubMed: 30927328
- DOI: https://doi.org/10.1002/pro.3610
- Primary Citation of Related Structures:
6IR0 - PubMed Abstract:
The Deltex (DTX) family is involved in ubiquitination and acts as Notch signaling modifiers for controlling cell fate determination. DTX promotes the development of the ubiquitin chain via its RING finger (DTX_RING). In this study, the solution structure of DTX_RING was determined using nuclear magnetic resonance (NMR). Moreover, by experiments with a metallochromic indicator, we spectrophotometrically estimated the stoichiometry of zinc ions and found that DTX_RING possesses zinc-binding capabilities. The Simple Modular Architecture Research Tool database predicted the structure of DTX_RING as a typical RING finger. However, the actual DTX_RING structure adopts a novel RING fold with a unique topology distinct from other RING fingers. We unveiled the position and the range of the DTX_RING active site at the atomic level. Artificial RING fingers (ARFs) are made by grafting active sites of the RING fingers onto cross-brace structure motifs. Therefore, the present structural analysis could be useful for designing a novel ARF.
Organizational Affiliation:
Department of Pharmaceutical Health Care, Faculty of Pharmaceutical Sciences, Himeji Dokkyo University, Himeji, Hyogo, Japan.