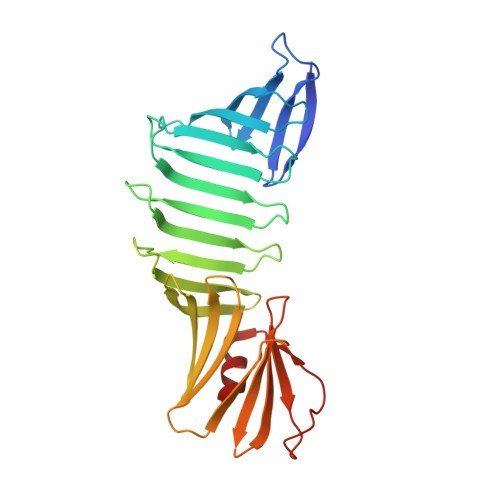
Domain-Swapping Design by Polyproline Rod Insertion.
Shiga, S., Yamanaka, M., Fujiwara, W., Hirota, S., Goda, S., Makabe, K.(2019) Chembiochem 20: 2454-2457
- PubMed: 31094059
- DOI: https://doi.org/10.1002/cbic.201900179
- Primary Citation of Related Structures:
6AIS, 6ICS, 6IDC, 6IEI, 6IYS - PubMed Abstract:
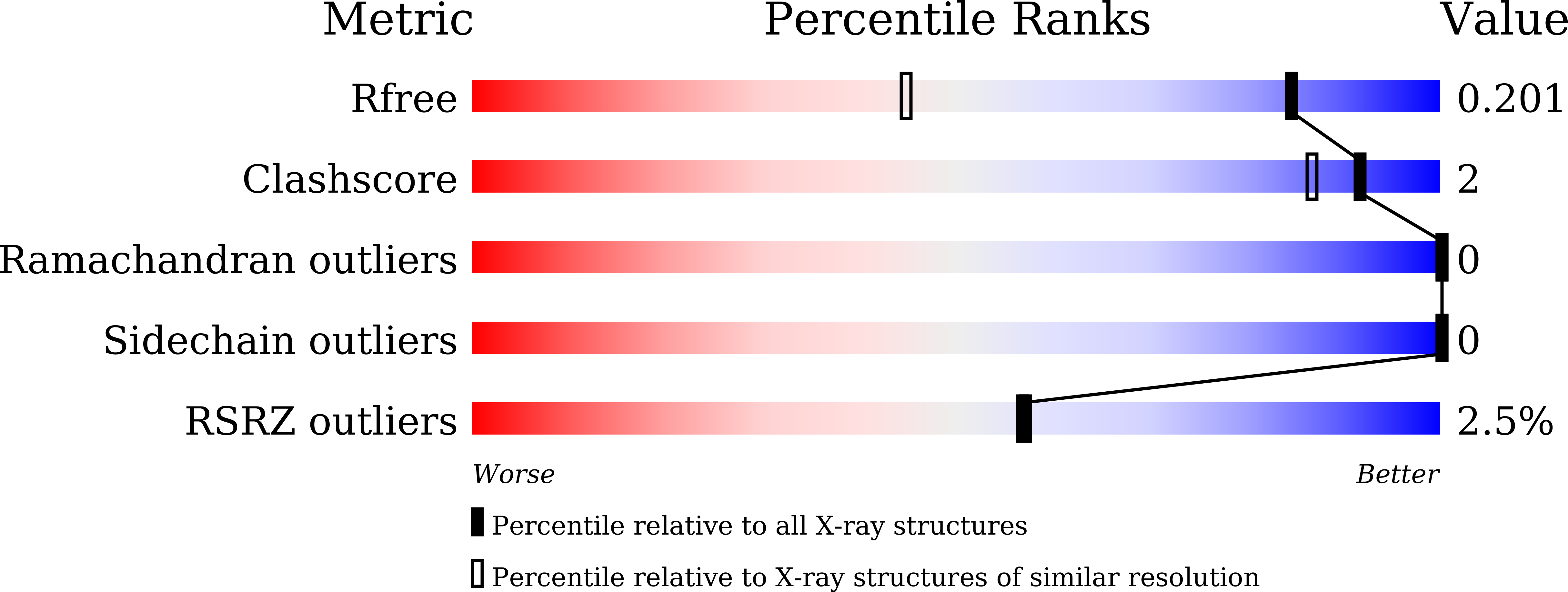
During domain swapping, proteins mutually interconvert structural elements to form a di-/oligomer. Engineering this process by design is important for creating a higher order protein assembly with minimal modification. Herein, a simple design strategy is shown for domain-swapping formation by loop deletion and insertion of a polyproline rod. Crystal structures revealed the formation of the domain-swapped dimers and polyproline portion formed a polyproline II (PPII) structure. Small-angle X-ray scattering demonstrated that an extended orientation of domain-swapped dimer was retained in solution. It is found that a multiple of three of inserting proline residue is favored for domain swapping because of the helical nature of PPII. The rigid nature of the polyproline rod enables precise control of the interdomain distance and orientation.
Organizational Affiliation:
Graduate School of Science and Engineering, Yamagata University, Jyonan 4-3-16, Yonezawa, Yamagata, 992-8510, Japan.