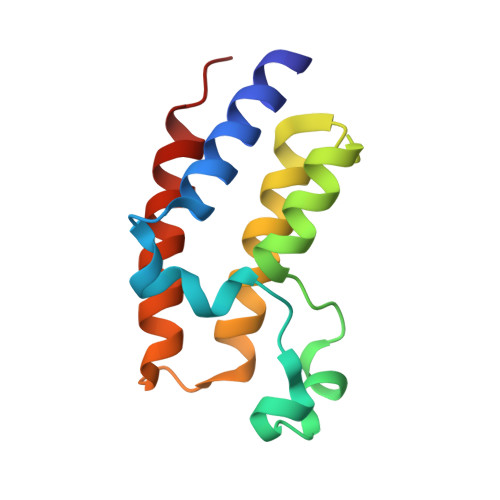
Crystal Structure of the second bromodomain of BRD2 in complex with RT53
Picaud, S., Traquete, R., Bernardes, G.J.L., Newman, J., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Filippakopoulos, P., Structural Genomics Consortium (SGC)To be published.