Global biochemical and structural analysis of the type IV pilus from the Gram-positive bacteriumStreptococcus sanguinis.
Berry, J.L., Gurung, I., Anonsen, J.H., Spielman, I., Harper, E., Hall, A.M.J., Goosens, V.J., Raynaud, C., Koomey, M., Biais, N., Matthews, S., Pelicic, V.(2019) J Biol Chem 294: 6796-6808
- PubMed: 30837269
- DOI: https://doi.org/10.1074/jbc.RA118.006917
- Primary Citation of Related Structures:
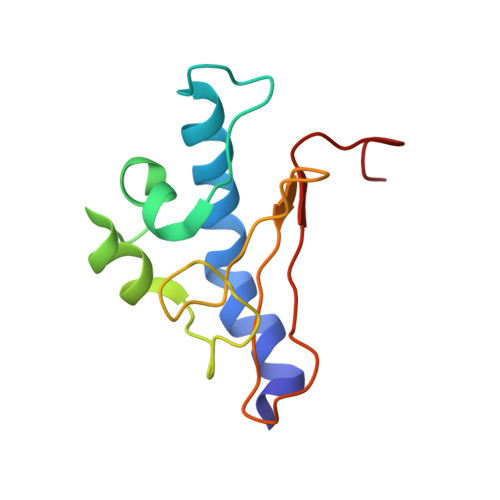
6I2O - PubMed Abstract:
Type IV pili (Tfp) are functionally versatile filaments, widespread in prokaryotes, that belong to a large class of filamentous nanomachines known as type IV filaments (Tff). Although Tfp have been extensively studied in several Gram-negative pathogens where they function as key virulence factors, many aspects of their biology remain poorly understood. Here, we performed a global biochemical and structural analysis of Tfp in a recently emerged Gram-positive model, Streptococcus sanguinis In particular, we focused on the five pilins and pilin-like proteins involved in Tfp biology in S. sanguinis We found that the two major pilins, PilE1 and PilE2, (i) follow widely conserved principles for processing by the prepilin peptidase PilD and for assembly into filaments; (ii) display only one of the post-translational modifications frequently found in pilins, i.e. a methylated N terminus; (iii) are found in the same heteropolymeric filaments; and (iv) are not functionally equivalent. The 3D structure of PilE1, solved by NMR, revealed a classical pilin-fold with a highly unusual flexible C terminus. Intriguingly, PilE1 more closely resembles pseudopilins forming shorter Tff than bona fide Tfp-forming major pilins, underlining the evolutionary relatedness among different Tff. Finally, we show that S. sanguinis Tfp contain a low abundance of three additional proteins processed by PilD, the minor pilins PilA, PilB, and PilC. These findings provide the first global biochemical and structural picture of a Gram-positive Tfp and have fundamental implications for our understanding of a widespread class of filamentous nanomachines.
Organizational Affiliation:
From the Medical Research Council Centre for Molecular Bacteriology and Infection, Imperial College London, London SW7 2AZ, United Kingdom.