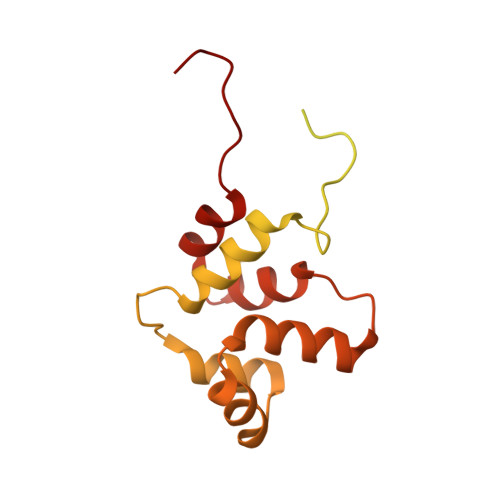
NMR structure of a full-length single-pass membrane protein NRADD.
Nadezhdin, K.D., Goncharuk, S.A., Arseniev, A.S., Mineev, K.S.(2019) Proteins 87: 786-790
- PubMed: 31033000
- DOI: https://doi.org/10.1002/prot.25703
- Primary Citation of Related Structures:
6HJ7 - PubMed Abstract:
Structural study of any single-pass membrane protein is both an important and challenging task. In this report, we present the structure of a neurotrophin receptor-alike death-domain protein. The structure and dynamics of the protein was investigated by conventional nuclear magnetic resonance techniques in the solution of phospholipid bicelles. The receptor contains two folded regions-α-helical transmembrane domain and globular C-terminal death domain with more than 50% of the rest of backbone being disordered. This is the first structure of a full-length single-pass membrane receptor-alike protein solved by the single method.
Organizational Affiliation:
Laboratory of biomolecular NMR spectroscopy, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia.