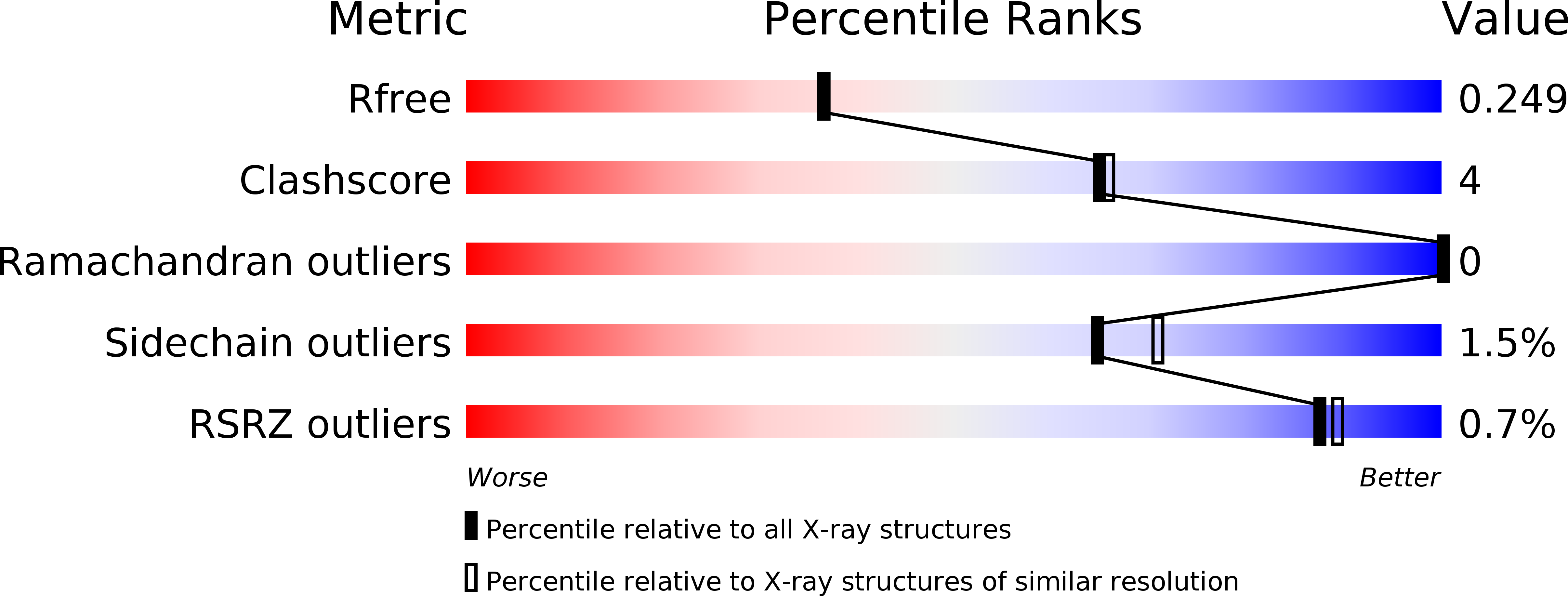
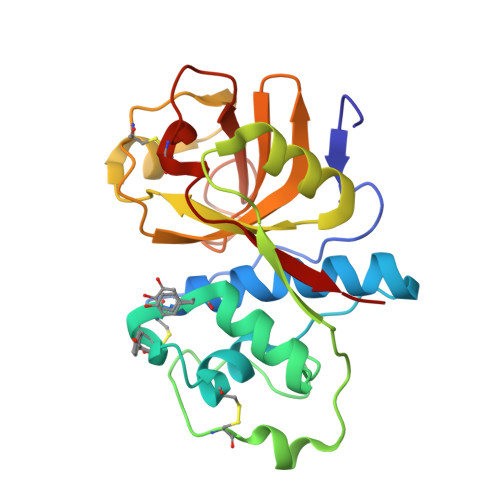
Crystallographic evidence for unexpected selective tyrosine hydroxylations in an aerated achiral Ru-papain conjugate.
Cherrier, M.V., Amara, P., Talbi, B., Salmain, M., Fontecilla-Camps, J.C.(2018) Metallomics 10: 1452-1459
- PubMed: 30175357
- DOI: https://doi.org/10.1039/c8mt00160j
- Primary Citation of Related Structures:
6H8T - PubMed Abstract:
The X-ray structure of an aerated achiral Ru-papain conjugate has revealed the hydroxylation of two tyrosine residues found near the ruthenium ion. The most likely mechanism involves a ruthenium-bound superoxide as the reactive species responsible for the first hydroxylation and the resulting high valent Ru(iv)[double bond, length as m-dash]O species for the second one.
Organizational Affiliation:
Univ. Grenoble Alpes, CEA, CNRS, IBS, Metalloproteins, F-38000 Grenoble, France. juan.fontecilla@ibs.fr.