Caged noble metals: Encapsulation of a cytotoxic platinum(II)-gold(I) compound within the ferritin nanocage.
Ferraro, G., Petruk, G., Maiore, L., Pane, F., Amoresano, A., Cinellu, M.A., Monti, D.M., Merlino, A.(2018) Int J Biol Macromol 115: 1116-1121
- PubMed: 29709536
- DOI: https://doi.org/10.1016/j.ijbiomac.2018.04.142
- Primary Citation of Related Structures:
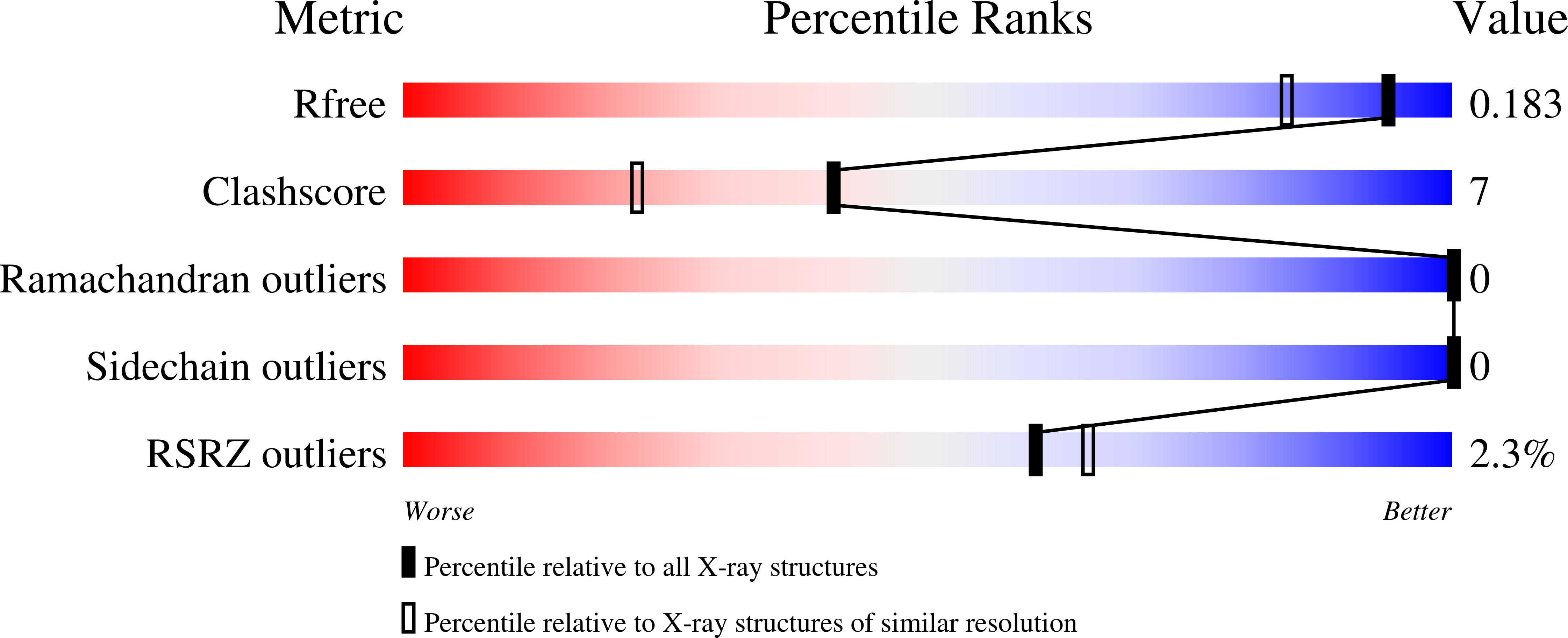
6FX8, 6FX9 - PubMed Abstract:
The encapsulation of Pt and Au-based anticancer agents within a protein cage is a promising way to enhance the selectivity of these potential drugs. Here a cytotoxic organometallic compound containing platinum(II) and gold(I) has been encapsulated within a ferritin nanocage (AFt). Inductively plasma coupled mass spectrometry data, collected to evaluate the amount of Pt and Au within the cage, indicate disruption of the starting heterobimetallic complex upon encapsulation within the nanocage. The drug-loaded protein (Pt(II)/Au(I)-AFt) has been characterized by UV-Vis spectroscopy, circular dichroism and X-ray diffraction analysis. Data indicate that the protein maintains its fold upon encapsulation of the metallodrug and that Au(I) and Pt(II)-containing fragments are encapsulated within the AFt cage, with Au(I) ion that binds the side chain of Cys126 and Pt(II) in the bulk, respectively. The in vitro cytotoxicity of Pt(II)Au(I)-AFt, as well as that of the free heterobimetallic complex, has been comparatively evaluated on human cervix and breast cancer cells and against cardiomyoblasts and keratinocytes non-tumorigenic cells. Our data demonstrate that it is possible to obtain a protein nanocarrier containing both Pt and Au atoms starting from a bimetallic compound, opening the way for the design and development of new potential drugs based on protein nanocarriers.
Organizational Affiliation:
Department of Chemical Sciences, University of Naples Federico II, Napoli, Italy.