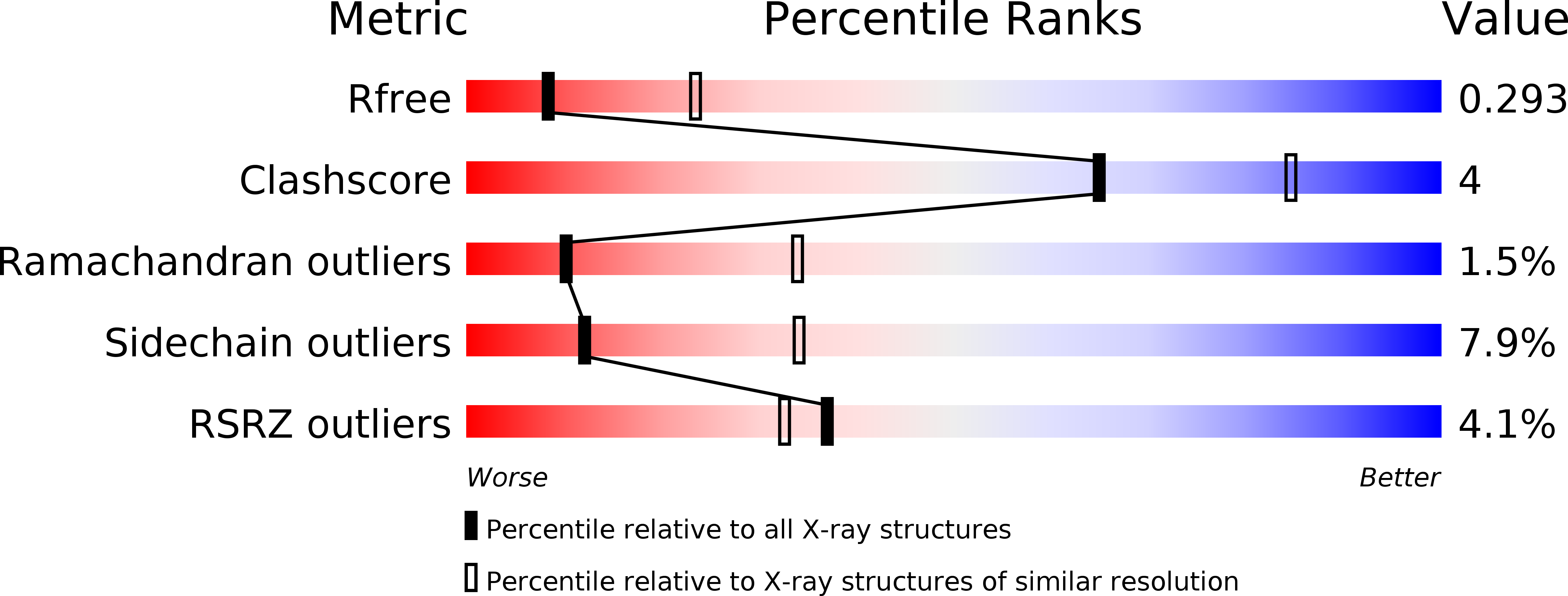
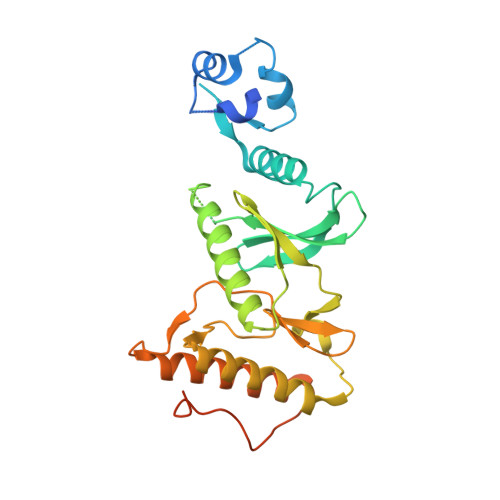
In vitro dimerization of human RIO2 kinase.
Maurice, F., Perebaskine, N., Thore, S., Fribourg, S.(2019) RNA Biol 16: 1633-1642
- PubMed: 31390939
- DOI: https://doi.org/10.1080/15476286.2019.1653679
- Primary Citation of Related Structures:
6FDM, 6FDN, 6FDO - PubMed Abstract:
RIO proteins form a conserved family of atypical protein kinases. RIO2 is a serine/threonine protein kinase/ATPase involved in pre-40S ribosomal maturation. Current crystal structures of archaeal and fungal Rio2 proteins report a monomeric form of the protein. Here, we describe three atomic structures of the human RIO2 kinase showing that it forms a homodimer in vitro . Upon self-association, each protomer ATP-binding pocket is partially remodelled and found in an apostate. The homodimerization is mediated by key residues previously shown to be responsible for ATP binding and catalysis. This unusual in vitro protein kinase dimer reveals an intricate mechanism where identical residues are involved in substrate binding and oligomeric state formation. We speculate that such an oligomeric state might be formed also in vivo and might function in maintaining the protein in an inactive state and could be employed during import.
Organizational Affiliation:
INSERM U1212, UMR CNRS 5320, Université de Bordeaux , Bordeaux , France.