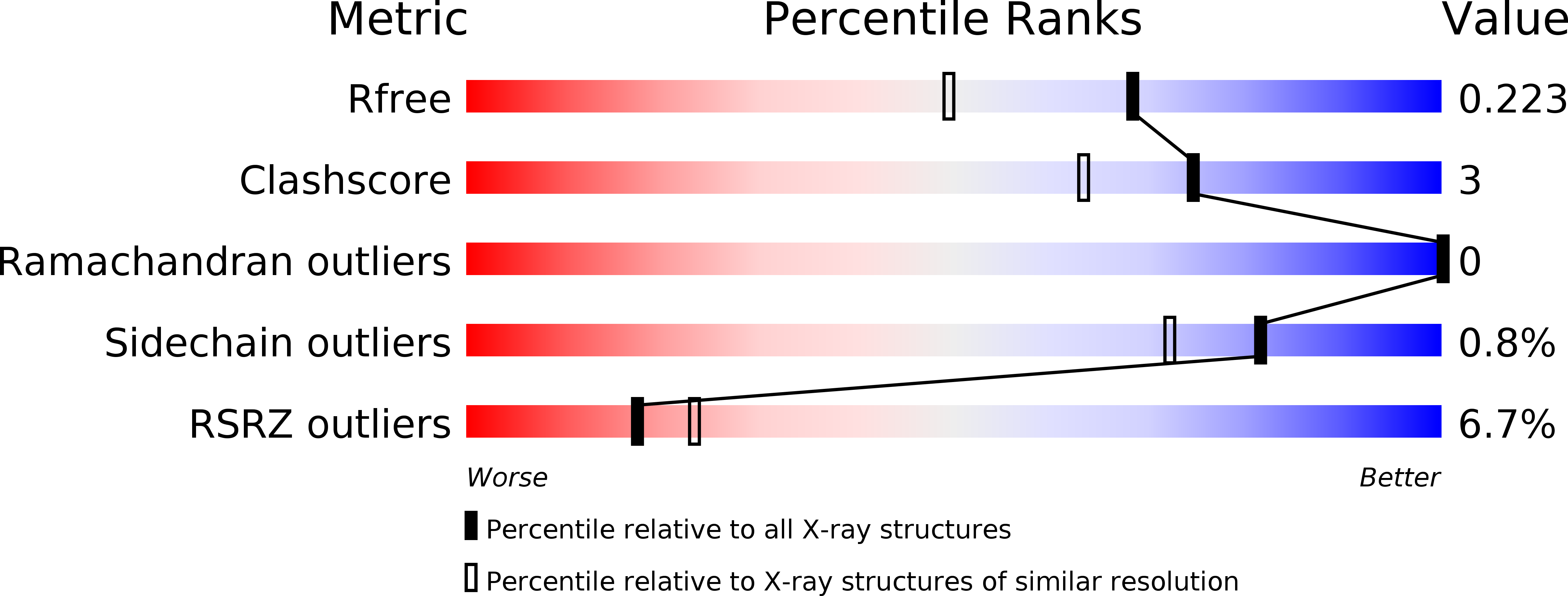
Human MARF1 is an endoribonuclease that interacts with the DCP1:2 decapping complex and degrades target mRNAs.
Nishimura, T., Fakim, H., Brandmann, T., Youn, J.Y., Gingras, A.C., Jinek, M., Fabian, M.R.(2018) Nucleic Acids Res 46: 12008-12021
- PubMed: 30364987
- DOI: https://doi.org/10.1093/nar/gky1011
- Primary Citation of Related Structures:
6FDL - PubMed Abstract:
Meiosis arrest female 1 (MARF1) is a cytoplasmic RNA binding protein that is essential for meiotic progression of mouse oocytes, in part by limiting retrotransposon expression. MARF1 is also expressed in somatic cells and tissues; however, its mechanism of action has yet to be investigated. Human MARF1 contains a NYN-like domain, two RRMs and eight LOTUS domains. Here we provide evidence that MARF1 post-transcriptionally silences targeted mRNAs. MARF1 physically interacts with the DCP1:DCP2 mRNA decapping complex but not with deadenylation machineries. Importantly, we provide a 1.7 Å resolution crystal structure of the human MARF1 NYN domain, which we demonstrate is a bona fide endoribonuclease, the activity of which is essential for the repression of MARF1-targeted mRNAs. Thus, MARF1 post-transcriptionally represses gene expression by serving as both an endoribonuclease and as a platform that recruits the DCP1:DCP2 decapping complex to targeted mRNAs.
Organizational Affiliation:
Lady Davis Institute for Medical Research, Jewish General Hospital, Montreal, Quebec, Canada.