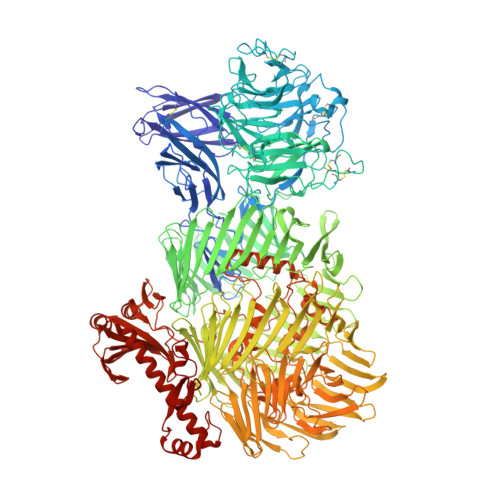
Structures of Teneurin adhesion receptors reveal an ancient fold for cell-cell interaction.
Jackson, V.A., Meijer, D.H., Carrasquero, M., van Bezouwen, L.S., Lowe, E.D., Kleanthous, C., Janssen, B.J.C., Seiradake, E.(2018) Nat Commun 9: 1079-1079
- PubMed: 29540701
- DOI: https://doi.org/10.1038/s41467-018-03460-0
- Primary Citation of Related Structures:
6FAY, 6FB3 - PubMed Abstract:
Teneurins are ancient cell-cell adhesion receptors that are vital for brain development and synapse organisation. They originated in early metazoan evolution through a horizontal gene transfer event when a bacterial YD-repeat toxin fused to a eukaryotic receptor. We present X-ray crystallography and cryo-EM structures of two Teneurins, revealing a ~200 kDa extracellular super-fold in which eight sub-domains form an intricate structure centred on a spiralling YD-repeat shell. An alternatively spliced loop, which is implicated in homophilic Teneurin interaction and specificity, is exposed and thus poised for interaction. The N-terminal side of the shell is 'plugged' via a fibronectin-plug domain combination, which defines a new class of YD proteins. Unexpectedly, we find that these proteins are widespread amongst modern bacteria, suggesting early metazoan receptor evolution from a distinct class of proteins, which today includes both bacterial proteins and eukaryotic Teneurins.
Organizational Affiliation:
Department of Biochemistry, Oxford University, OX1 3QU, Oxford, UK. verity.jackson@magd.ox.ac.uk.