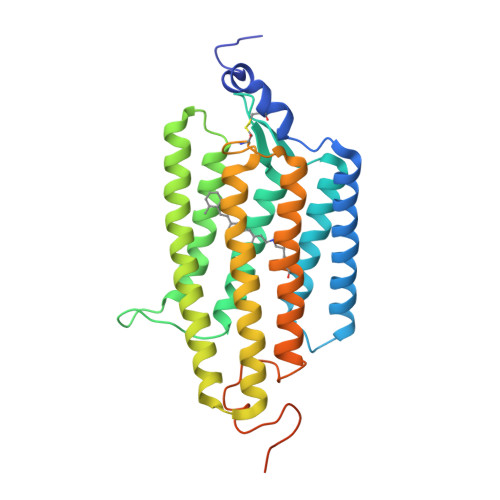
Crystal structure of a natural light-gated anion channelrhodopsin.
Li, H., Huang, C.Y., Govorunova, E.G., Schafer, C.T., Sineshchekov, O.A., Wang, M., Zheng, L., Spudich, J.L.(2019) Elife 8
- PubMed: 30614787
- DOI: https://doi.org/10.7554/eLife.41741
- Primary Citation of Related Structures:
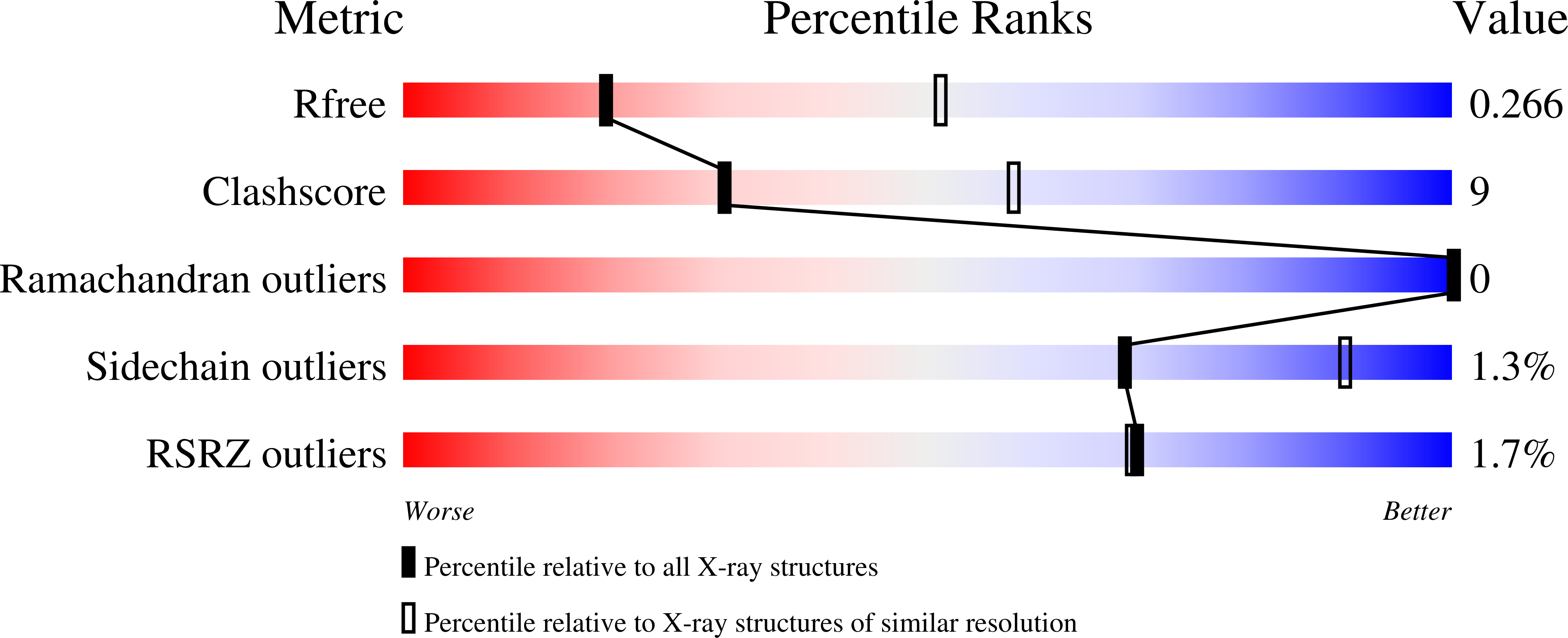
6EDQ - PubMed Abstract:
The anion channelrhodopsin Gt ACR1 from the alga Guillardia theta is a potent neuron-inhibiting optogenetics tool. Presented here, its X-ray structure at 2.9 Å reveals a tunnel traversing the protein from its extracellular surface to a large cytoplasmic cavity. The tunnel is lined primarily by small polar and aliphatic residues essential for anion conductance. A disulfide-immobilized extracellular cap facilitates channel closing and the ion path is blocked mid-membrane by its photoactive retinylidene chromophore and further by a cytoplasmic side constriction. The structure also reveals a novel photoactive site configuration that maintains the retinylidene Schiff base protonated when the channel is open. These findings suggest a new channelrhodopsin mechanism, in which the Schiff base not only controls gating, but also serves as a direct mediator for anion flux.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Center for Membrane Biology, University of Texas Health Science Center - McGovern Medical School, Houston, United States.