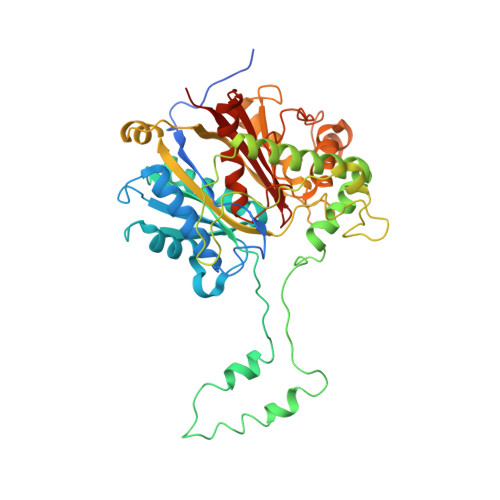
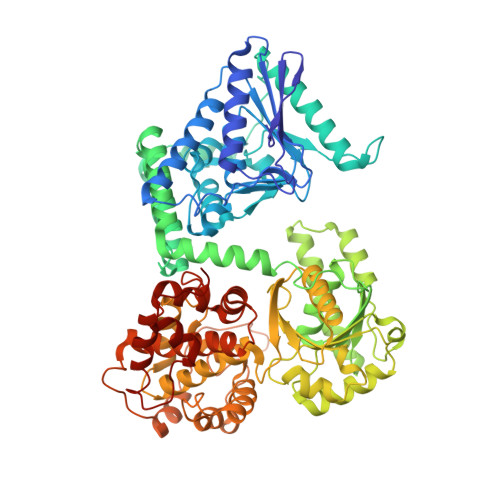
Crystal structure of human mitochondrial trifunctional protein, a fatty acid beta-oxidation metabolon.
Xia, C., Fu, Z., Battaile, K.P., Kim, J.P.(2019) Proc Natl Acad Sci U S A 116: 6069-6074
- PubMed: 30850536
- DOI: https://doi.org/10.1073/pnas.1816317116
- Primary Citation of Related Structures:
6DV2 - PubMed Abstract:
Membrane-bound mitochondrial trifunctional protein (TFP) catalyzes β-oxidation of long chain fatty acyl-CoAs, employing 2-enoyl-CoA hydratase (ECH), 3-hydroxyl-CoA dehydrogenase (HAD), and 3-ketothiolase (KT) activities consecutively. Inherited deficiency of TFP is a recessive genetic disease, manifesting in hypoketotic hypoglycemia, cardiomyopathy, and sudden death. We have determined the crystal structure of human TFP at 3.6-Å resolution. The biological unit of the protein is α 2 β 2 The overall structure of the heterotetramer is the same as that observed by cryo-EM methods. The two β-subunits make a tightly bound homodimer at the center, and two α-subunits are bound to each side of the β 2 dimer, creating an arc, which binds on its concave side to the mitochondrial innermembrane. The catalytic residues in all three active sites are arranged similarly to those of the corresponding, soluble monofunctional enzymes. A structure-based, substrate channeling pathway from the ECH active site to the HAD and KT sites is proposed. The passage from the ECH site to the HAD site is similar to those found in the two bacterial TFPs. However, the passage from the HAD site to the KT site is unique in that the acyl-CoA intermediate can be transferred between the two sites by passing along the mitochondrial inner membrane using the hydrophobic nature of the acyl chain. The 3'-AMP-PPi moiety is guided by the positively charged residues located along the "ceiling" of the channel, suggesting that membrane integrity is an essential part of the channel and is required for the activity of the enzyme.
Organizational Affiliation:
Department of Biochemistry, Medical College of Wisconsin, Milwaukee, WI 53226.