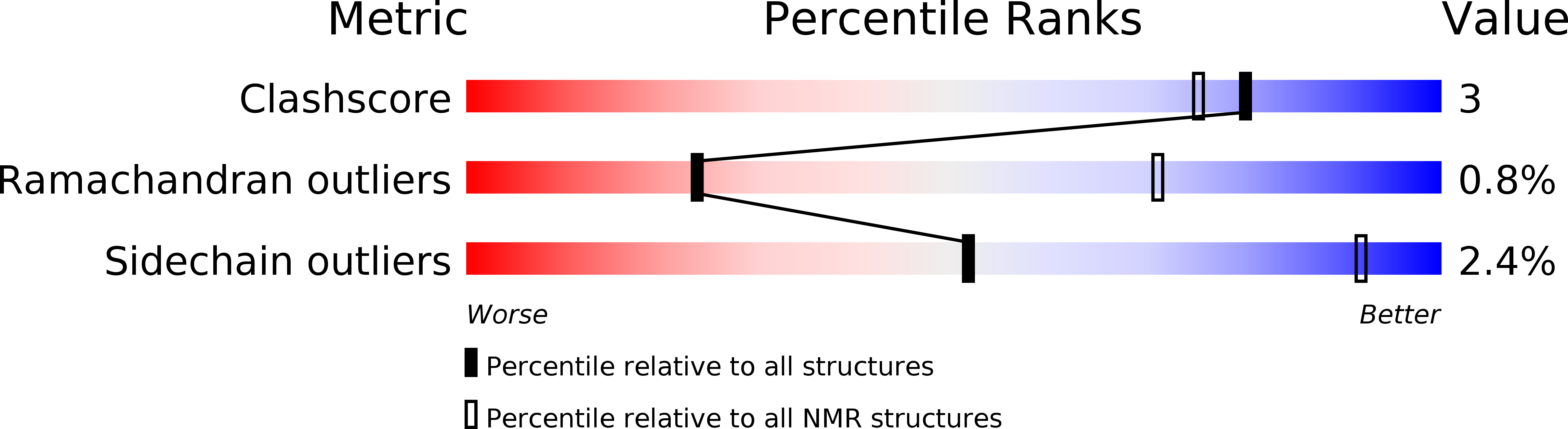An Atypical Mechanism of Split Intein Molecular Recognition and Folding.
Stevens, A.J., Sekar, G., Gramespacher, J.A., Cowburn, D., Muir, T.W.(2018) J Am Chem Soc 140: 11791-11799
- PubMed: 30156841
- DOI: https://doi.org/10.1021/jacs.8b07334
- Primary Citation of Related Structures:
6DSL - PubMed Abstract:
Split inteins associate to trigger protein splicing in trans, a post-translational modification in which protein sequences fused to the intein pair are ligated together in a traceless manner. Recently, a family of naturally split inteins has been identified that is split at a noncanonical location in the primary sequence. These atypically split inteins show considerable promise in protein engineering applications; however, the mechanism by which they associate is unclear and must be different from that of previously characterized canonically split inteins due to unique topological restrictions. Here, we use a consensus design strategy to generate an atypical split intein pair (Cat) that has greatly improved activity and is amenable to detailed biochemical and biophysical analysis. Guided by the solution structure of Cat, we show that the association of the fragments involves a disorder-to-order structural transition driven by hydrophobic interactions. This molecular recognition mechanism satisfies the topological constraints of the intein fold and, importantly, ensures that premature chemistry does not occur prior to fragment complementation. Our data lead a common blueprint for split intein complementation in which localized structural rearrangements are used to drive folding and regulate protein-splicing activity.
Organizational Affiliation:
Department of Chemistry, Frick Laboratory , Princeton University , Princeton , New Jersey 08544 , United States.