Protein Scaffold Activates Catalytic CO2 Hydrogenation by a Rhodium Bis(diphosphine) Complex
Laureanti, J.A., Buckho, G.W., Katipamula, S., Su, Q., Linehan, J.C., Zadvornyy, O.A., Peters, J.W., O'Hagan, M.(2019) ACS Catal 9: 620-625
Experimental Data Snapshot
(2019) ACS Catal 9: 620-625
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Transcriptional regulator, PadR-like family | 117 | Lactococcus cremoris subsp. cremoris MG1363 | Mutation(s): 3 Gene Names: llmg_0323 | 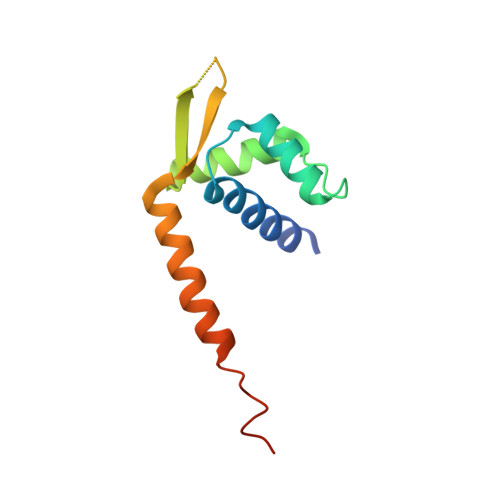 | |
UniProt | |||||
Find proteins for A2RI36 (Lactococcus lactis subsp. cremoris (strain MG1363)) Explore A2RI36 Go to UniProtKB: A2RI36 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A2RI36 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| JY1 Query on JY1 | C [auth A] | bis[diethyl(methyl)-lambda~5~-phosphanyl]{bis[{[(2-{[2-(2,5-dioxopyrrolidin-1-yl)ethyl]amino}-2-oxoethyl)amino]methyl}(diethyl)-lambda~5~-phosphanyl]}rhodium C36 H74 N6 O6 P4 Rh WBSWUPZUVFVXQZ-UHFFFAOYSA-R |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 101.645 | α = 90 |
| b = 35.569 | β = 96.51 |
| c = 71.259 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Department of Energy (DOE, United States) | United States | DE-SC0012518 |
| Other government | United States | PNNL Laboratory Directed Research and Development 69937 |