Noncanonical role for the binding protein in substrate uptake by the MetNI methionine ATP Binding Cassette (ABC) transporter.
Nguyen, P.T., Lai, J.Y., Lee, A.T., Kaiser, J.T., Rees, D.C.(2018) Proc Natl Acad Sci U S A 115: E10596-E10604
- PubMed: 30352853
- DOI: https://doi.org/10.1073/pnas.1811003115
- Primary Citation of Related Structures:
6CVL - PubMed Abstract:
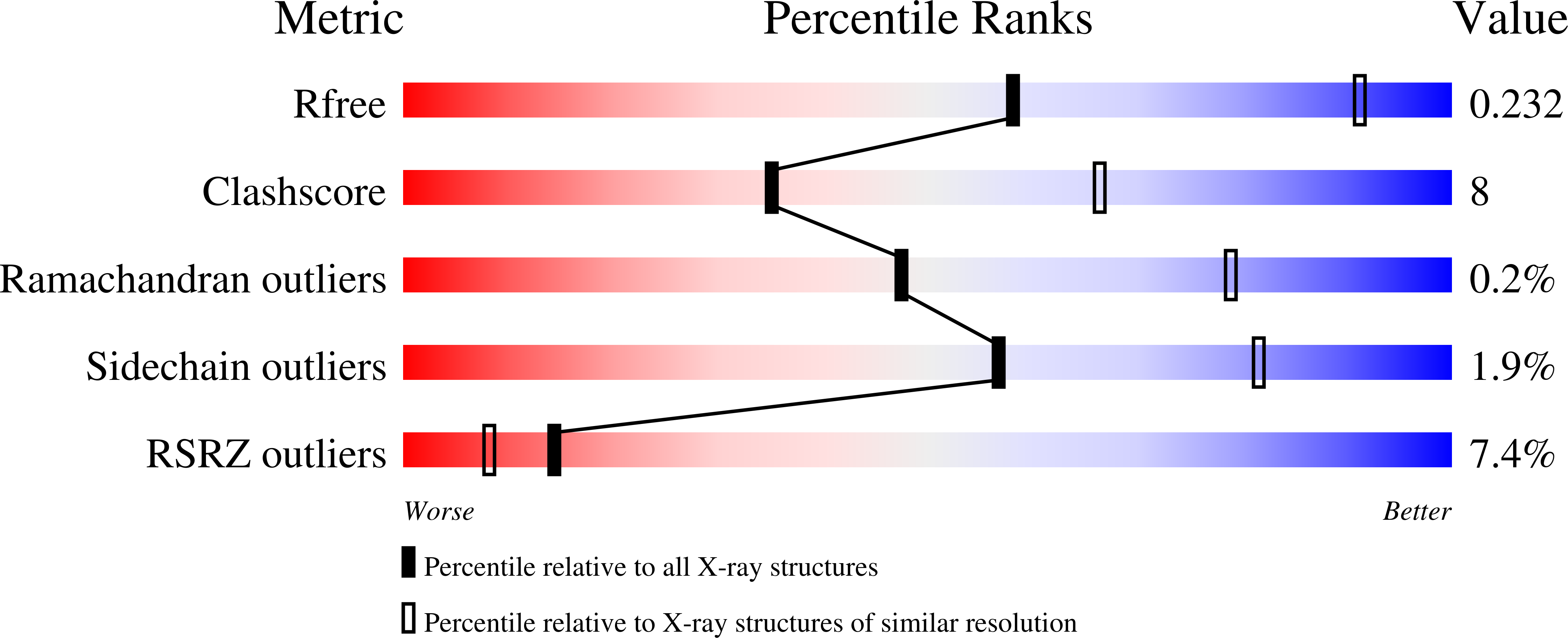
The Escherichia coli methionine ABC transporter MetNI exhibits both high-affinity transport toward l-methionine and broad specificity toward methionine derivatives, including d-methionine. In this work, we characterize the transport of d-methionine derivatives by the MetNI transporter. Unexpectedly, the N229A substrate-binding deficient variant of the cognate binding protein MetQ was found to support high MetNI transport activity toward d-selenomethionine. We determined the crystal structure at 2.95 Å resolution of the ATPγS-bound MetNIQ complex in the outward-facing conformation with the N229A apo MetQ variant. This structure revealed conformational changes in MetQ providing substrate access through the binding protein to the transmembrane translocation pathway. MetQ likely mediates uptake of methionine derivatives through two mechanisms: in the methionine-bound form delivering substrate from the periplasm to the transporter (the canonical mechanism) and in the apo form by facilitating ligand binding when complexed to the transporter (the noncanonical mechanism). This dual role for substrate-binding proteins is proposed to provide a kinetic strategy for ABC transporters to transport both high- and low-affinity substrates present in a physiological concentration range.
Organizational Affiliation:
Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA 91125.