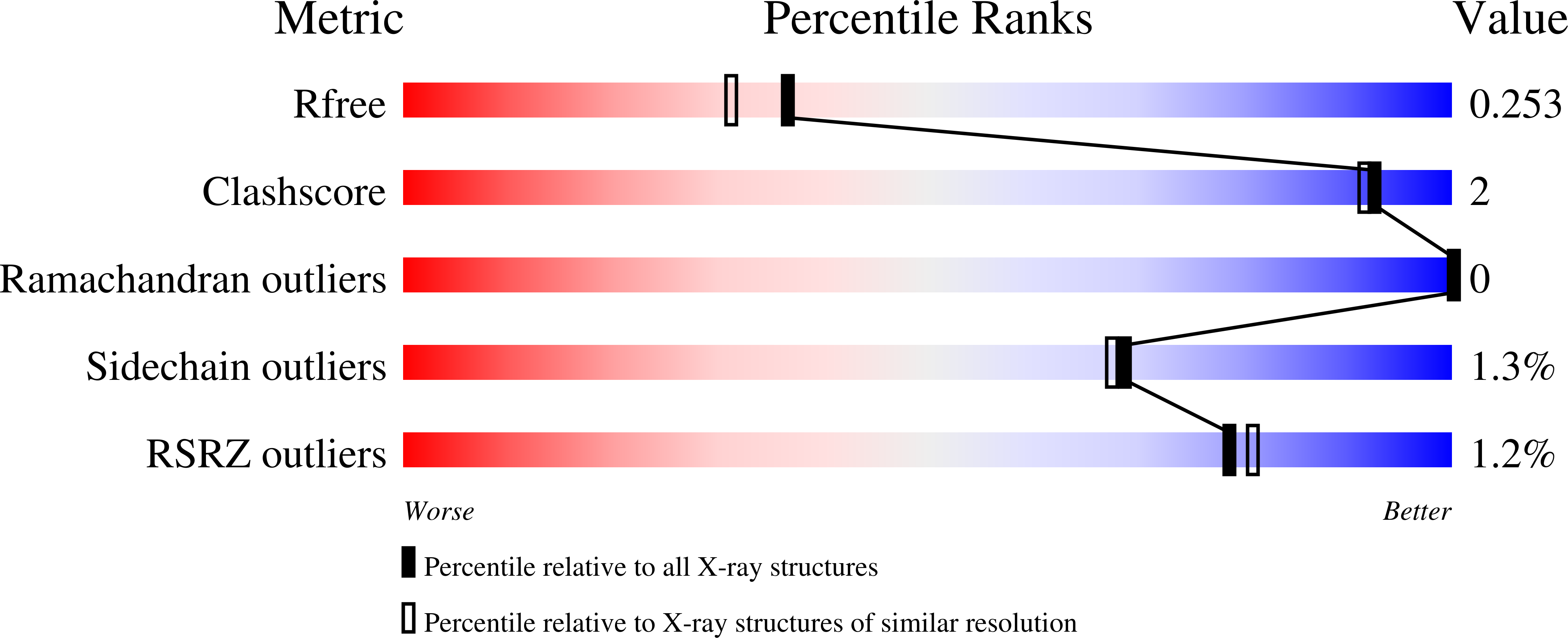
Mouse IgG2c Fc loop residues promote greater receptor-binding affinity than mouse IgG2b or human IgG1.
Falconer, D.J., Barb, A.W.(2018) PLoS One 13: e0192123-e0192123
- PubMed: 29408873
- DOI: https://doi.org/10.1371/journal.pone.0192123
- Primary Citation of Related Structures:
6BHQ, 6BHY - PubMed Abstract:
The structures of non-human antibodies are largely unstudied despite the potential for the identification of alternative structural motifs and physical properties that will benefit a basic understanding of protein and immune system evolution as well as highlight unexplored motifs to improve therapeutic monoclonal antibody. Here we probe the structure and receptor-binding properties of the mouse IgG2c crystallizable fragment (Fc) to compare to mouse IgG2b and human IgG1 Fcs. Models of mIgG2c Fc determined by x-ray crystallography with a complex-type biantennary (to 2.05 Å) or a truncated (1)GlcNAc asparagine-linked (N)-glycan attached (to 2.04 Å) show differences in key regions related to mouse Fc γ receptor IV (mFcγRIV) binding. Mouse IgG2c forms different non-bonded interactions between the BC, DE and FG loops than the highly-conserved mIgG2b and binds to FcγRIV with 4.7-fold greater affinity in the complex-type glycoform. Secondary structural elements surrounding the Asn297 site of glycosylation form longer beta strands in the truncated mIgG2c Fc glycoform when compared to mIgG2c with the complex-type N-glycan. Solution NMR spectroscopy of the N-linked (1)GlcNAc residues show differences between mIgG2b, 2c and hIgG1 Fc that correlate to receptor binding affinity. Mutations targeting differences in mIgG2 DE and FG loops decreased affinity of mIgG2c for FcγRIV and increased affinity of mIgG2b. Changes in NMR spectra of the mutant Fc proteins mirrored these changes in affinity. Our studies identified structural and functional differences in highly conserved molecules that were not predicted from primary sequence comparison.
Organizational Affiliation:
Roy J. Carver Department of Biochemistry, Biophysics and Molecular Biology Iowa State University, Ames, IA, United States of America.