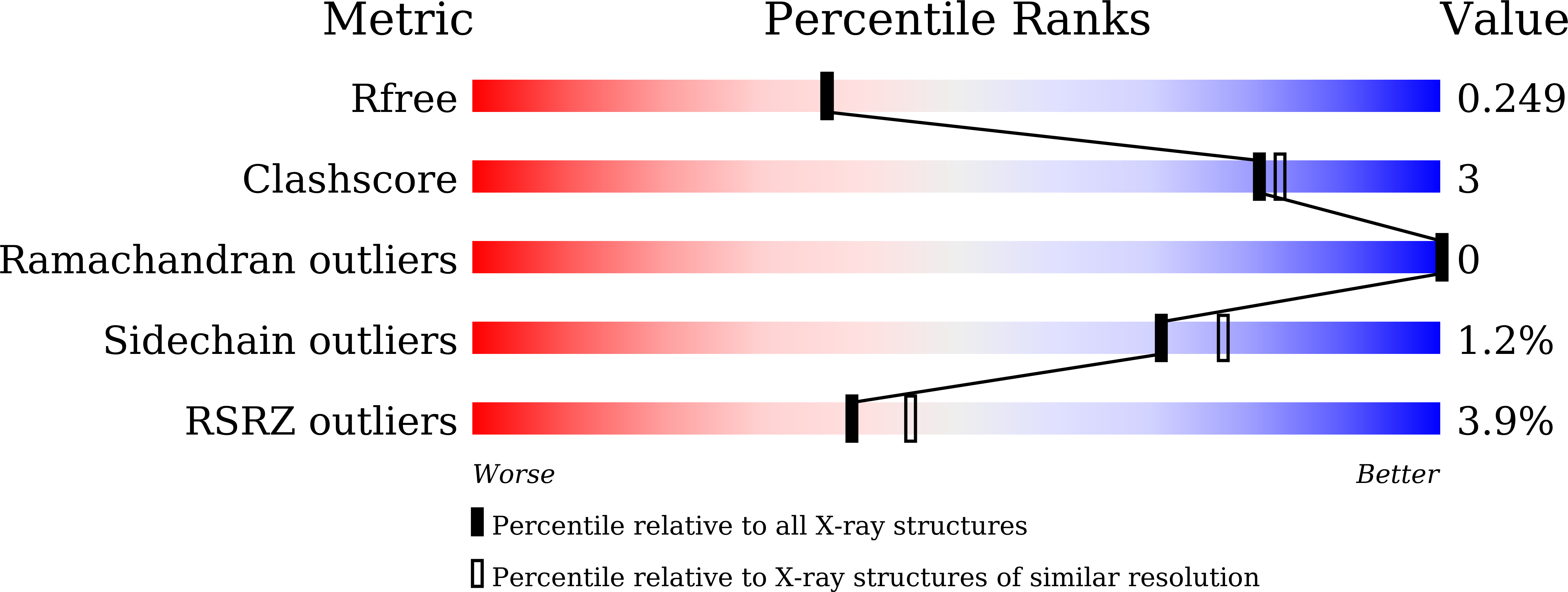
Crystallographic Analysis of the Catalytic Mechanism of Phosphopantothenoylcysteine Synthetase from Saccharomyces cerevisiae.
Zheng, P., Zhang, M., Khan, M.H., Liu, H., Jin, Y., Yue, J., Gao, Y., Teng, M., Zhu, Z., Niu, L.(2019) J Mol Biol 431: 764-776
- PubMed: 30653991
- DOI: https://doi.org/10.1016/j.jmb.2019.01.012
- Primary Citation of Related Structures:
6AI8, 6AI9, 6AIK, 6AIM, 6AIP - PubMed Abstract:
Phosphopantothenoylcysteine (PPC) synthetase (PPCS) catalyzes nucleoside triphosphate-dependent condensation reaction between 4'-phosphopantothenate (PPA) and l-cysteine to form PPC in CoA biosynthesis. The catalytic mechanism of PPCS has not been resolved yet. Coenzyme A biosynthesis protein 2 (Cab2) possesses activity of PPCS in Saccharomyces cerevisiae. Our enzymatic assays suggest that Cab2 could utilize both ATP and CTP to activate PPA in vitro. The results of isothermal titration calorimetry indicate that PPA, CTP, and ATP could bind to Cab2 individually, with PPA having the highest binding affinity. To provide further insight into the catalytic mechanism of Cab2, we determined the crystal structures of Cab2 and its complex with PPA, the reaction intermediate 4'-phosphopantothenoyl-CMP, the final reaction product PPC, and the product analogue phosphopantothenoylcystine. Except for PPA, all other ligands were generated in situ and present in the active-site pocket of Cab2. Structures of Cab2 in complex with ligands provide insight into substrates binding and its catalytic mechanism. Analysis of structures indicates that the carboxyl of PPA-moiety of ligands and the γ-amino group of Asn97 possess different conformations in these complex structures. The cysteine/cystine/serine selectivity assays for Cab2 indicate that the amino group rather than the thiol group of l-cysteine attacks the carbonyl of 4'-phosphopantothenoyl-CMP to form PPC. Based on structural and biochemical data, the catalytic mechanism of Cab2 was proposed for the first time.
Organizational Affiliation:
Hefei National Laboratory for Physical Sciences at Microscale, School of Life Sciences, University of Science and Technology of China, 96 Jinzhai Road, Hefei, Anhui 230026, China.